Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro
- PMID: 34835024
- PMCID: PMC8621278
- DOI: 10.3390/v13112218
Stephan Oroszlan and the Proteolytic Processing of Retroviral Proteins: Following A Pro
Abstract
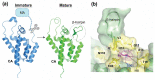
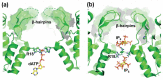
Steve Oroszlan determined the sequences at the ends of virion proteins for a number of different retroviruses. This work led to the insight that the amino-terminal amino acid of the mature viral CA protein is always proline. In this remembrance, we review Steve's work that led to this insight and show how that insight was a necessary precursor to the work we have done in the subsequent years exploring the cleavage rate determinants of viral protease processing sites and the multiple roles the amino-terminal proline of CA plays after protease cleavage liberates it from its position in a protease processing site.
Keywords: HIV-1; Oroszlan; capsid; protease; retroviruses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Personal name as subject
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

