Curing Cats with Feline Infectious Peritonitis with an Oral Multi-Component Drug Containing GS-441524
- PMID: 34835034
- PMCID: PMC8621566
- DOI: 10.3390/v13112228
Curing Cats with Feline Infectious Peritonitis with an Oral Multi-Component Drug Containing GS-441524
Abstract
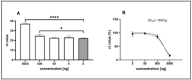
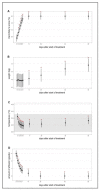
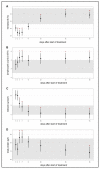
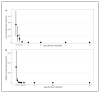
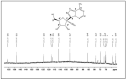
Feline infectious peritonitis (FIP) caused by feline coronavirus (FCoV) is a common dis-ease in cats, fatal if untreated, and no effective treatment is currently legally available. The aim of this study was to evaluate efficacy and toxicity of the multi-component drug Xraphconn® in vitro and as oral treatment in cats with spontaneous FIP by examining survival rate, development of clinical and laboratory parameters, viral loads, anti-FCoV antibodies, and adverse effects. Mass spectrometry and nuclear magnetic resonance identified GS-441524 as an active component of Xraphconn®. Eighteen cats with FIP were prospectively followed up while being treated orally for 84 days. Values of key parameters on each examination day were compared to values before treatment initiation using linear mixed-effect models. Xraphconn® displayed high virucidal activity in cell culture. All cats recovered with dramatic improvement of clinical and laboratory parameters and massive reduction in viral loads within the first few days of treatment without serious adverse effects. Oral treatment with Xraphconn® containing GS-441524 was highly effective for FIP without causing serious adverse effects. This drug is an excellent option for the oral treatment of FIP and should be trialed as potential effective treatment option for other severe coronavirus-associated diseases across species.
Keywords: FCoV; FIP; GS-441524; Mutian; Xraphconn®; antiviral chemotherapy; feline coronavirus; therapy; treatment.
Conflict of interest statement
The authors declare that they have no conflict of interest. The oral multi-component drug Xraphconn® was provided by Mutian Life Sciences Limited, but Mutian played no role in the interpretation of study data or the decision to submit the manuscript for publication. No commercial conflict of interest exists as the information is solely for scientific dissemination.
Figures



References
-
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

