CRISPR Screening: Molecular Tools for Studying Virus-Host Interactions
- PMID: 34835064
- PMCID: PMC8618713
- DOI: 10.3390/v13112258
CRISPR Screening: Molecular Tools for Studying Virus-Host Interactions
Abstract
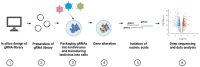
CRISPR/Cas is a powerful tool for studying the role of genes in viral infections. The invention of CRISPR screening technologies has made it possible to untangle complex interactions between the host and viral agents. Moreover, whole-genome and pathway-specific CRISPR screens have facilitated identification of novel drug candidates for treating viral infections. In this review, we highlight recent developments in the fields of CRISPR/Cas with a focus on the use of CRISPR screens for studying viral infections and identifying new candidate genes to aid development of antivirals.
Keywords: COVID-19; Cas12; Cas9; HAV; HIV; SARS-CoV-2; ZIKV; hepatitis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

