Rift Valley Fever Virus Propagates in Human Villous Trophoblast Cell Lines and Induces Cytokine mRNA Responses Known to Provoke Miscarriage
- PMID: 34835071
- PMCID: PMC8625252
- DOI: 10.3390/v13112265
Rift Valley Fever Virus Propagates in Human Villous Trophoblast Cell Lines and Induces Cytokine mRNA Responses Known to Provoke Miscarriage
Abstract
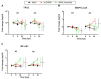
The mosquito-borne Rift Valley fever (RVF) is a prioritised disease that has been listed by the World Health Organization for urgent research and development of counteraction. Rift Valley fever virus (RVFV) can cause a cytopathogenic effect in the infected cell and induce hyperimmune responses that contribute to pathogenesis. In livestock, the consequences of RVFV infection vary from mild symptoms to abortion. In humans, 1-3% of patients with RVFV infection develop severe disease, manifested as, for example, haemorrhagic fever, encephalitis or blindness. RVFV infection has also been associated with miscarriage in humans. During pregnancy, there should be a balance between pro-inflammatory and anti-inflammatory mediators to create a protective environment for the placenta and foetus. Many viruses are capable of penetrating that protective environment and infecting the foetal-maternal unit, possibly via the trophoblasts in the placenta, with potentially severe consequences. Whether it is the viral infection per se, the immune response, or both that contribute to the pathogenesis of miscarriage remains unknown. To investigate how RVFV could contribute to pathogenesis during pregnancy, we infected two human trophoblast cell lines, A3 and Jar, representing normal and transformed human villous trophoblasts, respectively. They were infected with two RVFV variants (wild-type RVFV and RVFV with a deleted NSs protein), and the infection kinetics and 15 different cytokines were analysed. The trophoblast cell lines were infected by both RVFV variants and infection caused upregulation of messenger RNA (mRNA) expression for interferon (IFN) types I-III and inflammatory cytokines, combined with cell line-specific mRNA expression of transforming growth factor (TGF)-β1 and interleukin (IL)-10. When comparing the two RVFV variants, we found that infection with RVFV lacking NSs function caused a hyper-IFN response and inflammatory response, while the wild-type RVFV suppressed the IFN I and inflammatory response. The induction of certain cytokines by RVFV infection could potentially lead to teratogenic effects that disrupt foetal and placental developmental pathways, leading to birth defects and other pregnancy complications, such as miscarriage.
Keywords: cytokine; human villous trophoblast; inflammatory cytokines; interferon; miscarriage; rift valley fever virus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
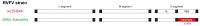


References
-
- World Health Organization. [(accessed on 14 August 2020)]. Available online: https://www.who.int/activities/prioritizing-diseases-for-research-and-de....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

