Huntingtin-Interacting Protein 1 Promotes Vpr-Induced G2 Arrest and HIV-1 Infection in Macrophages
- PMID: 34835114
- PMCID: PMC8624357
- DOI: 10.3390/v13112308
Huntingtin-Interacting Protein 1 Promotes Vpr-Induced G2 Arrest and HIV-1 Infection in Macrophages
Abstract
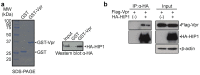
Human immunodeficiency virus type 1 (HIV-1) modulates the host cell cycle. The HIV-1 accessory protein Vpr arrests the cell cycle at the G2 phase in dividing cells, and the ability of Vpr to induce G2 arrest is well conserved among primate lentiviruses. Additionally, Vpr-mediated G2 arrest likely correlates with enhanced HIV-1 infection in monocyte-derived macrophages. Here, we screened small-interfering RNA to reveal candidates that suppress Vpr-induced G2 arrest and identified Huntingtin-interacting protein 1 (HIP1) required for efficient G2 arrest. Interestingly, HIP1 was not essential for Vpr-induced DNA double-strand breaks, which are required for activation of the DNA-damage checkpoint and G2 arrest. Furthermore, HIP1 knockdown suppressed HIV-1 infection in monocyte-derived macrophages. This study identifies HIP1 as a factor promoting Vpr-induced G2 arrest and HIV-1 infection in macrophages.
Keywords: G2 arrest; HIP1; HIV-1; Vpr; macrophage.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



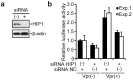
Similar articles
-
Human Immunodeficiency Virus Type 1 Vpr Mediates Degradation of APC1, a Scaffolding Component of the Anaphase-Promoting Complex/Cyclosome.J Virol. 2021 Jul 12;95(15):e0097120. doi: 10.1128/JVI.00971-20. Epub 2021 Jul 12. J Virol. 2021. PMID: 34011540 Free PMC article.
-
HIV-1 Vpr Protein Enhances Proteasomal Degradation of MCM10 DNA Replication Factor through the Cul4-DDB1[VprBP] E3 Ubiquitin Ligase to Induce G2/M Cell Cycle Arrest.J Biol Chem. 2015 Jul 10;290(28):17380-9. doi: 10.1074/jbc.M115.641522. Epub 2015 Jun 1. J Biol Chem. 2015. PMID: 26032416 Free PMC article.
-
HIV-1 Vpr-induced DNA damage activates NF-κB through ATM-NEMO independent of cell cycle arrest.mBio. 2024 Oct 16;15(10):e0024024. doi: 10.1128/mbio.00240-24. Epub 2024 Sep 13. mBio. 2024. PMID: 39269169 Free PMC article.
-
HIV-1 Vpr: G2 cell cycle arrest, macrophages and nuclear transport.Prog Cell Cycle Res. 1997;3:21-7. doi: 10.1007/978-1-4615-5371-7_2. Prog Cell Cycle Res. 1997. PMID: 9552403 Review.
-
Defining the roles for Vpr in HIV-1-associated neuropathogenesis.J Neurovirol. 2016 Aug;22(4):403-15. doi: 10.1007/s13365-016-0436-5. Epub 2016 Apr 7. J Neurovirol. 2016. PMID: 27056720 Free PMC article. Review.
Cited by
-
HIV-1 Vpr Functions in Primary CD4+ T Cells.Viruses. 2024 Mar 9;16(3):420. doi: 10.3390/v16030420. Viruses. 2024. PMID: 38543785 Free PMC article. Review.
-
The innate immune factor RPRD2/REAF and its role in the Lv2 restriction of HIV.mBio. 2023 Dec 19;14(6):e0257221. doi: 10.1128/mbio.02572-21. Epub 2023 Oct 26. mBio. 2023. PMID: 37882563 Free PMC article. Review.
-
Genotoxic consequences of viral infections.Npj Viruses. 2025 Jan 27;3(1):5. doi: 10.1038/s44298-024-00087-5. Npj Viruses. 2025. PMID: 40295867 Free PMC article. Review.
-
A Novel Class of HIV-1 Inhibitors Targeting the Vpr-Induced G2-Arrest in Macrophages by New Yeast- and Cell-Based High-Throughput Screening.Viruses. 2022 Jun 16;14(6):1321. doi: 10.3390/v14061321. Viruses. 2022. PMID: 35746791 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

