Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview
- PMID: 34835248
- PMCID: PMC8622998
- DOI: 10.3390/vaccines9111317
Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview
Abstract
Respiratory viruses represent a major public health concern, as they are highly mutated, resulting in new strains emerging with high pathogenicity. Currently, the world is suffering from the newly evolving severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus is the cause of coronavirus disease 2019 (COVID-19), a mild-to-severe respiratory tract infection with frequent ability to give rise to fatal pneumonia in humans. The overwhelming outbreak of SARS-CoV-2 continues to unfold all over the world, urging scientists to put an end to this global pandemic through biological and pharmaceutical interventions. Currently, there is no specific treatment option that is capable of COVID-19 pandemic eradication, so several repurposed drugs and newly conditionally approved vaccines are in use and heavily applied to control the COVID-19 pandemic. The emergence of new variants of the virus that partially or totally escape from the immune response elicited by the approved vaccines requires continuous monitoring of the emerging variants to update the content of the developed vaccines or modify them totally to match the new variants. Herein, we discuss the potential therapeutic and prophylactic interventions including repurposed drugs and the newly developed/approved vaccines, highlighting the impact of virus evolution on the immune evasion of the virus from currently licensed vaccines for COVID-19.
Keywords: COVID-19; SARS-CoV-2; clinical trials; drug repurposing; management; vaccines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
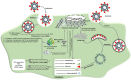
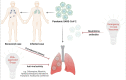
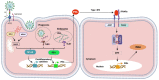
References
-
- El Gizawy H.A., Boshra S.A., Mostafa A., Mahmoud S.H., Ismail M.I., Alsfouk A.A., Taher A.T., Al-Karmalawy A.A. Pimenta dioica (L.) Merr. Bioactive Constituents Exert Anti-SARS-CoV-2 and Anti-Inflammatory Activities: Molecular Docking and Dynamics, In Vitro, and In Vivo Studies. Molecules. 2021;26:5844. doi: 10.3390/molecules26195844. - DOI - PMC - PubMed
-
- King A.M., Lefkowitz E., Adams M.J., Carstens E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Volume 9 Elsevier; Amsterdam, The Netherlands: 2011.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

