Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii
- PMID: 34835462
- PMCID: PMC8619257
- DOI: 10.3390/microorganisms9112338
Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii
Abstract
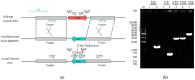
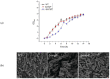
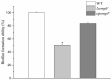
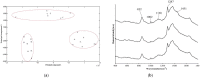
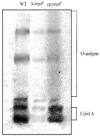
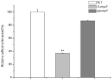
In some Gram-negative bacteria, ompF encodes outer membrane protein F (OmpF), which is a cation-selective porin and is responsible for the passive transport of small molecules across the outer membrane. However, there are few reports about the functions of this gene in Cronobacter sakazakii. To investigate the role of ompF in detail, an ompF disruption strain (ΔompF) and a complementation strain (cpompF) were successfully obtained. We find that OmpF can affect the ability of biofilm formation in C. sakazakii. In addition, the variations in biofilm composition of C. sakazakii were examined using Raman spectroscopy analyses caused by knocking out ompF, and the result indicated that the levels of certain biofilm components, including lipopolysaccharide (LPS), were significantly decreased in the mutant (ΔompF). Then, SDS-PAGE was used to further analyze the LPS content, and the result showed that the LPS levels were significantly reduced in the absence of ompF. Therefore, we conclude that OmpF affects biofilm formation in C. sakazakii by reducing the amount of LPS. Furthermore, the ΔompF mutant showed decreased (2.7-fold) adhesion to and invasion of HCT-8 cells. In an antibiotic susceptibility analysis, the ΔompF mutant showed significantly smaller inhibition zones than the WT, indicating that OmpF had a positive effect on the influx of antibiotics into the cells. In summary, ompF plays a positive regulatory role in the biofilm formation and adhesion/invasion, which is achieved by regulating the amount of LPS, but is a negative regulator of antibiotic resistance in C. sakazakii.
Keywords: Cronobacter sakazakii; LPS; adhesion/invasion; biofilm formation; ompF.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

