Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe
- PMID: 34835519
- PMCID: PMC8617893
- DOI: 10.3390/microorganisms9112394
Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe
Abstract
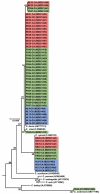
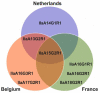
Cryptosporidium is an apicomplexan parasitic protist, which infects a wide range of hosts, causing cryptosporidiosis disease. In farms, the incidence of this disease is high in animals such as cows, leading to extensive economic loss in the livestock industry. Infected cows may also act as a major reservoir of Cryptosporidium spp., in particular C. parvum, the most common cause of cryptosporidiosis in these animals. This poses a risk to the trading of livestock, to other farms via breeding centres, and to human health. This study is a part of a global project aimed at strategies to tackle cryptosporidiosis. To reach this target, it was essential to determine whether prevalence was dependent on the studied countries or if the issue was borderless. Indeed, C. parvum occurrence was assessed across dairy farms in certain regions of Belgium, France, and the Netherlands. At the same time, the animal-to-animal transmission of the circulating C. parvum subtypes was studied. To accomplish this, we analysed 1084 faecal samples, corresponding to 57 dairy farms from all three countries. To this end, 18S rRNA and gp60 genes fragments were amplified, followed by DNA sequencing, which was subsequently used for detection and subtyping C. parvum. Bioinformatic and phylogenetic methods were integrated to analyse and characterise the obtained DNA sequences. Our results show 25.7%, 24.9% and 20.8% prevalence of Cryptosporidium spp. in Belgium, France, and the Netherlands respectively. Overall, 93% of the farms were Cryptosporidium positive. The gp60 subtyping demonstrated a significant number of the C. parvum positives belonged to the IIa allelic family, which has been also identified in humans. Therefore, this study highlights how prevalent C. parvum is in dairy farms and further suggests cattle as a possible carrier of zoonotic C. parvum subtypes, which could pose a threat to human health.
Keywords: 18S rRNA; Cryptosporidium; dairy cattle; genotyping; gp60; prevalence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Molecular characterization of Cryptosporidium isolates from diarrheal dairy calves in France.Vet Parasitol Reg Stud Reports. 2019 Dec;18:100323. doi: 10.1016/j.vprsr.2019.100323. Epub 2019 Jul 24. Vet Parasitol Reg Stud Reports. 2019. PMID: 31796198 Free PMC article.
-
Follow-up investigation into Cryptosporidium prevalence and transmission in Western European dairy farms.Vet Parasitol. 2023 Jun;318:109920. doi: 10.1016/j.vetpar.2023.109920. Epub 2023 Apr 1. Vet Parasitol. 2023. PMID: 37030025
-
Cryptosporidium parvum subtypes from diarrheic dairy calves in Israel.Vet Parasitol Reg Stud Reports. 2021 Jul;25:100608. doi: 10.1016/j.vprsr.2021.100608. Epub 2021 Jul 14. Vet Parasitol Reg Stud Reports. 2021. PMID: 34474801
-
Cryptosporidium parvum and gp60 genotype prevalence in dairy calves worldwide: a systematic review and meta-analysis.Acta Trop. 2023 Apr;240:106843. doi: 10.1016/j.actatropica.2023.106843. Epub 2023 Feb 3. Acta Trop. 2023. PMID: 36738819
-
Emergence of zoonotic Cryptosporidium parvum in China.Trends Parasitol. 2022 Apr;38(4):335-343. doi: 10.1016/j.pt.2021.12.002. Epub 2021 Dec 28. Trends Parasitol. 2022. PMID: 34972653 Review.
Cited by
-
Cryptosporidium parvum: an emerging occupational zoonosis in Finland.Acta Vet Scand. 2023 Jun 22;65(1):25. doi: 10.1186/s13028-023-00684-z. Acta Vet Scand. 2023. PMID: 37349848 Free PMC article.
-
The cow GUTBIOME CY study: investigating the composition of the cattle gut microbiome in health and infectious disease transmission in cyprus.BMC Vet Res. 2024 Dec 19;20(1):566. doi: 10.1186/s12917-024-04419-8. BMC Vet Res. 2024. PMID: 39696220 Free PMC article.
-
First Epidemiological Report on the Prevalence and Associated Risk Factors of Cryptosporidium spp. in Farmed Marine and Wild Freshwater Fish in Central and Eastern of Algeria.Acta Parasitol. 2022 Sep;67(3):1152-1161. doi: 10.1007/s11686-022-00560-2. Epub 2022 May 11. Acta Parasitol. 2022. PMID: 35545736
-
The Evaluation of the Efficacy of a Novel Subunit Vaccine in the Prevention of Cryptosporidium parvum-Caused Diarrhoea in Neonatal Calves.Animals (Basel). 2025 Jan 8;15(2):132. doi: 10.3390/ani15020132. Animals (Basel). 2025. PMID: 39858132 Free PMC article.
-
Study of the economic impact of cryptosporidiosis in calves after implementing good practices to manage the disease on dairy farms in Belgium, France, and the Netherlands.Curr Res Parasitol Vector Borne Dis. 2023 Oct 10;4:100149. doi: 10.1016/j.crpvbd.2023.100149. eCollection 2023. Curr Res Parasitol Vector Borne Dis. 2023. PMID: 37941926 Free PMC article.
References
-
- Checkley W., White A.C., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X.M., Fayer R., Griffiths J.K., Guerrant R.L., Hedstrom L., et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

