Identifying the Predictive Role of Oxidative Stress Genes in the Prognosis of Glioma Patients
- PMID: 34836934
- PMCID: PMC8634738
- DOI: 10.12659/MSM.934161
Identifying the Predictive Role of Oxidative Stress Genes in the Prognosis of Glioma Patients
Abstract
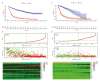
BACKGROUND Gliomas are primary aggressive brain tumors with poor prognoses. Oxidative stress plays a crucial role in the tumorigenesis and drug resistance of gliomas. The aim of the present study was to use integrated bioinformatics analyses to evaluate the prognostic value of oxidative stress-related genes (OSRGs) in glioma. MATERIAL AND METHODS Disease- and prognosis-associated OSRGs were identified using microarray and clinical data from the Chinese Glioma Genome Atlas database. Functional enrichment, gene-gene interaction, protein-protein interaction, and survival analyses were performed in screened OSRGs. The protein expression was validated by the Human Protein Atlas database. A risk score model was constructed and verified through Cox regression, receiver operating characteristic curve, principal component, and stratified analyses. The Cancer Genome Atlas (TCGA) database was used for external validation. A nomogram was constructed to facilitate the clinical application. RESULTS Twenty-one disease-associated and 14 prognosis-associated OSRGs were identified. Enrichment analyses indicated that these signature OSRGs were involved in tumorigenesis and drug resistance of glioma. The risk score model demonstrated a significant difference in overall survival between the high- and low-risk groups. The area under the curve and hazard ratio (1.296) revealed the independent prognostic value of the model. The model exhibited good predictive efficacy in the TCGA cohort. A clinical nomogram was constructed to calculate survival rates in glioma patients at 1, 3, and 5 years. CONCLUSIONS Our comprehensive study indicated that OSRGs were valuable for prognosis prediction in glioma, which provides a novel insight into the relationship between oxidative stress and glioma and a potential therapeutic strategy for glioma patients.
Conflict of interest statement
Figures








References
-
- Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–46. - PubMed
-
- Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131(6):803–20. - PubMed
-
- Cairncross G, Jenkins R. Gliomas with 1p/19q codeletion: a.k.a. oligodendroglioma. Cancer J. 2008;14(6):352–57. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

