CRISPR-enhanced human adipocyte browning as cell therapy for metabolic disease
- PMID: 34836963
- PMCID: PMC8626495
- DOI: 10.1038/s41467-021-27190-y
CRISPR-enhanced human adipocyte browning as cell therapy for metabolic disease
Abstract
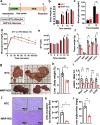
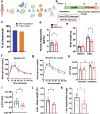
Obesity and type 2 diabetes are associated with disturbances in insulin-regulated glucose and lipid fluxes and severe comorbidities including cardiovascular disease and steatohepatitis. Whole body metabolism is regulated by lipid-storing white adipocytes as well as "brown" and "brite/beige" adipocytes that express thermogenic uncoupling protein 1 (UCP1) and secrete factors favorable to metabolic health. Implantation of brown fat into obese mice improves glucose tolerance, but translation to humans has been stymied by low abundance of primary human beige adipocytes. Here we apply methods to greatly expand human adipocyte progenitors from small samples of human subcutaneous adipose tissue and then disrupt the thermogenic suppressor gene NRIP1 by CRISPR. Ribonucleoprotein consisting of Cas9 and sgRNA delivered ex vivo are fully degraded by the human cells following high efficiency NRIP1 depletion without detectable off-target editing. Implantation of such CRISPR-enhanced human or mouse brown-like adipocytes into high fat diet fed mice decreases adiposity and liver triglycerides while enhancing glucose tolerance compared to implantation with unmodified adipocytes. These findings advance a therapeutic strategy to improve metabolic homeostasis through CRISPR-based genetic enhancement of human adipocytes without exposing the recipient to immunogenic Cas9 or delivery vectors.
© 2021. The Author(s).
Conflict of interest statement
The authors declare the following competing Interests: M.P.C. and A.J.G. are inventors of granted US Patent #8,519,118, “RIP140 regulation of glucose transport”, and of granted US Patent #8,093,223, “RIP140 regulation of diabetes”, related to data in this paper on Nrip1/RIP140-depleted mouse and human adipocytes. SC is inventor on granted US Patent #10,093,902, “Human adipose tissue white and brown-on-white progenitors for reconstructive and metabolic therapies”, related to data in this paper on human adipose progenitors and human adipocytes. M.P.C., S.C., E.T., and S.M.N. are inventors of pending US Patent application #63/089,955, “Targeting Nrip1 to Alleviate Metabolic Disease”, related to CRISPR-based depletion of Nrip1 in mouse and human adipocytes in this paper. The University of Massachusetts is the grantee or potential grantee of all of the above. MPC and SC declare that they are bound by confidentiality agreements that prevent them from disclosing a potentially competing interest in this work. E.J.S. is a co-founder and scientific advisor of Intellia Therapeutics. The remaining authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

