Epidemiological associations with genomic variation in SARS-CoV-2
- PMID: 34837008
- PMCID: PMC8626494
- DOI: 10.1038/s41598-021-02548-w
Epidemiological associations with genomic variation in SARS-CoV-2
Abstract
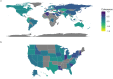
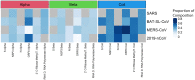
SARS-CoV-2 (CoV) is the etiological agent of the COVID-19 pandemic and evolves to evade both host immune systems and intervention strategies. We divided the CoV genome into 29 constituent regions and applied novel analytical approaches to identify associations between CoV genomic features and epidemiological metadata. Our results show that nonstructural protein 3 (nsp3) and Spike protein (S) have the highest variation and greatest correlation with the viral whole-genome variation. S protein variation is correlated with nsp3, nsp6, and 3'-to-5' exonuclease variation. Country of origin and time since the start of the pandemic were the most influential metadata associated with genomic variation, while host sex and age were the least influential. We define a novel statistic-coherence-and show its utility in identifying geographic regions (populations) with unusually high (many new variants) or low (isolated) viral phylogenetic diversity. Interestingly, at both global and regional scales, we identify geographic locations with high coherence neighboring regions of low coherence; this emphasizes the utility of this metric to inform public health measures for disease spread. Our results provide a direction to prioritize genes associated with outcome predictors (e.g., health, therapeutic, and vaccine outcomes) and to improve DNA tests for predicting disease status.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Ardeshirdavani, A. et al. Clinical population genetic analysis of variants in the SARS-CoV-2 receptor ACE2. medRxiv (2020).
-
- de Cruz JO, de Oliveira Cruz J, Sousa SMB. SARS-CoV-2 receptor and renin-angiotensin system regulation: Impact of genetics variants in ACE2 gene impact of genetics variants in the ACE2 gene in the functional receptor of SARS-CoV-2. Int. J. Innov. Sci. Res. Technol. 2020;5:489–497. doi: 10.38124/IJISRT20JUL268. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

