Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics
- PMID: 34837014
- PMCID: PMC9018519
- DOI: 10.1038/s41579-021-00660-2
Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics
Abstract
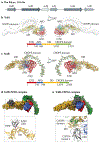
Clostridioides difficile is a Gram-positive anaerobe that can cause a spectrum of disorders that range in severity from mild diarrhoea to fulminant colitis and/or death. The bacterium produces up to three toxins, which are considered the major virulence factors in C. difficile infection. These toxins promote inflammation, tissue damage and diarrhoea. In this Review, we highlight recent biochemical and structural advances in our understanding of the mechanisms that govern host-toxin interactions. Understanding how C. difficile toxins affect the host forms a foundation for developing novel strategies for treatment and prevention of C. difficile infection.
© 2021. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
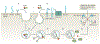


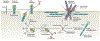

References
-
- CDC. Antibiotic Resistance Threats in the United States (CDC, 2019).
-
- Shen A Clostridioides difficile spore formation and germination: new insights and opportunities for intervention. Annu. Rev. Microbiol 74, 545–566 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

