Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses - are we our own worst enemy?
- PMID: 34837062
- PMCID: PMC8617551
- DOI: 10.1038/s41577-021-00656-2
Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses - are we our own worst enemy?
Erratum in
-
Author Correction: Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses - are we our own worst enemy?Nat Rev Immunol. 2022 Mar;22(3):200. doi: 10.1038/s41577-021-00673-1. Nat Rev Immunol. 2022. PMID: 34921295 Free PMC article. No abstract available.
Abstract
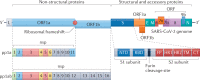
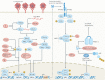
Human coronaviruses cause a wide spectrum of disease, ranging from mild common colds to acute respiratory distress syndrome and death. Three highly pathogenic human coronaviruses - severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus and SARS-CoV-2 - have illustrated the epidemic and pandemic potential of human coronaviruses, and a better understanding of their disease-causing mechanisms is urgently needed for the rational design of therapeutics. Analyses of patients have revealed marked dysregulation of the immune system in severe cases of human coronavirus infection, and there is ample evidence that aberrant immune responses to human coronaviruses are typified by impaired induction of interferons, exuberant inflammatory responses and delayed adaptive immune responses. In addition, various viral proteins have been shown to impair interferon induction and signalling and to induce inflammasome activation. This suggests that severe disease associated with human coronaviruses is mediated by both dysregulated host immune responses and active viral interference. Here we discuss our current understanding of the mechanisms involved in each of these scenarios.
© 2021. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
ORF9c and ORF10 as accessory proteins of SARS-CoV-2 in immune evasion.Nat Rev Immunol. 2022 May;22(5):331. doi: 10.1038/s41577-022-00715-2. Nat Rev Immunol. 2022. PMID: 35361899 Free PMC article. No abstract available.
References
-
- CDC. SARS. Basics Factsheet. https://www.cdc.gov/sars/about/fs-sars.html (2017).
-
- WHO. EMRO. MERS outbreaks. MERS-CoV. Health topics. http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html (2021).
-
- WHO. WHO coronavirus (COVID-19) dashboard. https://covid19.who.int. (2021).
-
- Schoggins JW. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 2019;6:567–584. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

