Deep learning enables genetic analysis of the human thoracic aorta
- PMID: 34837083
- PMCID: PMC8758523
- DOI: 10.1038/s41588-021-00962-4
Deep learning enables genetic analysis of the human thoracic aorta
Abstract
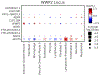
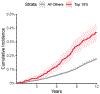
Enlargement or aneurysm of the aorta predisposes to dissection, an important cause of sudden death. We trained a deep learning model to evaluate the dimensions of the ascending and descending thoracic aorta in 4.6 million cardiac magnetic resonance images from the UK Biobank. We then conducted genome-wide association studies in 39,688 individuals, identifying 82 loci associated with ascending and 47 with descending thoracic aortic diameter, of which 14 loci overlapped. Transcriptome-wide analyses, rare-variant burden tests and human aortic single nucleus RNA sequencing prioritized genes including SVIL, which was strongly associated with descending aortic diameter. A polygenic score for ascending aortic diameter was associated with thoracic aortic aneurysm in 385,621 UK Biobank participants (hazard ratio = 1.43 per s.d., confidence interval 1.32-1.54, P = 3.3 × 10-20). Our results illustrate the potential for rapidly defining quantitative traits with deep learning, an approach that can be broadly applied to biomedical images.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures













References
-
- Benjamin EJ et al. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation 139, e56–e528 (2019). - PubMed
-
- Isselbacher EM Thoracic and abdominal aortic aneurysms. Circulation 111, 816–828 (2005). - PubMed
-
- Owens DK et al. Screening for abdominal aortic aneurysm: US Preventive Services Task Force Recommendation Statement. JAMA 322, 2211–2218 (2019). - PubMed
-
- Fann JI Descending thoracic and thoracoabdominal aortic aneurysms. Coron. Artery Dis 13, 93–102 (2002). - PubMed
-
- Guo D-C, Papke CL, He R & Milewicz DM Pathogenesis of thoracic and abdominal aortic aneurysms. Ann. N. Y. Acad. Sci 1085, 339–352 (2006). - PubMed
Methods-only References
-
- He K, Zhang X, Ren S & Sun J Deep residual learning for image recognition. ArXiv151203385 Cs (2015).
-
- Krizhevsky A, Sutskever I & Hinton GE ImageNet classification with deep convolutional neural networks. Communications of the ACM 60, 84–90 (2017).
Publication types
MeSH terms
Grants and funding
- K08 HL159346/HL/NHLBI NIH HHS/United States
- R01 HL139731/HL/NHLBI NIH HHS/United States
- R01 HL141434/HL/NHLBI NIH HHS/United States
- K01 HL140187/HL/NHLBI NIH HHS/United States
- T32 HL007208/HL/NHLBI NIH HHS/United States
- 18SFRN34250007/AHA/American Heart Association-American Stroke Association/United States
- K24 HL105780/HL/NHLBI NIH HHS/United States
- R01 HL140224/HL/NHLBI NIH HHS/United States
- R01 HL092577/HL/NHLBI NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- U54 HL120163/HL/NHLBI NIH HHS/United States
- R01 HL128914/HL/NHLBI NIH HHS/United States
- R01 HL157635/HL/NHLBI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- K24 HL153669/HL/NHLBI NIH HHS/United States
- R01 HL134893/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

