A graph-based gene selection method for medical diagnosis problems using a many-objective PSO algorithm
- PMID: 34838034
- PMCID: PMC8627636
- DOI: 10.1186/s12911-021-01696-3
A graph-based gene selection method for medical diagnosis problems using a many-objective PSO algorithm
Abstract
Background: Gene expression data play an important role in bioinformatics applications. Although there may be a large number of features in such data, they mainly tend to contain only a few samples. This can negatively impact the performance of data mining and machine learning algorithms. One of the most effective approaches to alleviate this problem is to use gene selection methods. The aim of gene selection is to reduce the dimensions (features) of gene expression data leading to eliminating irrelevant and redundant genes.
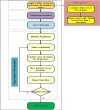
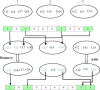
Methods: This paper presents a hybrid gene selection method based on graph theory and a many-objective particle swarm optimization (PSO) algorithm. To this end, a filter method is first utilized to reduce the initial space of the genes. Then, the gene space is represented as a graph to apply a graph clustering method to group the genes into several clusters. Moreover, the many-objective PSO algorithm is utilized to search an optimal subset of genes according to several criteria, which include classification error, node centrality, specificity, edge centrality, and the number of selected genes. A repair operator is proposed to cover the whole space of the genes and ensure that at least one gene is selected from each cluster. This leads to an increasement in the diversity of the selected genes.
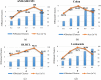
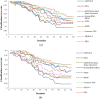
Results: To evaluate the performance of the proposed method, extensive experiments are conducted based on seven datasets and two evaluation measures. In addition, three classifiers-Decision Tree (DT), Support Vector Machine (SVM), and K-Nearest Neighbors (KNN)-are utilized to compare the effectiveness of the proposed gene selection method with other state-of-the-art methods. The results of these experiments demonstrate that our proposed method not only achieves more accurate classification, but also selects fewer genes than other methods.
Conclusion: This study shows that the proposed multi-objective PSO algorithm simultaneously removes irrelevant and redundant features using several different criteria. Also, the use of the clustering algorithm and the repair operator has improved the performance of the proposed method by covering the whole space of the problem.
Keywords: Dimension reduction; Gene clustering; Gene selection; High dimensional; Many-objective PSO; Repair operator.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
A comparative analysis of feature selection models for spatial analysis of floods using hybrid metaheuristic and machine learning models.Environ Sci Pollut Res Int. 2024 May;31(23):33495-33514. doi: 10.1007/s11356-024-33389-5. Epub 2024 Apr 29. Environ Sci Pollut Res Int. 2024. PMID: 38684613
-
A Tri-Stage Wrapper-Filter Feature Selection Framework for Disease Classification.Sensors (Basel). 2021 Aug 18;21(16):5571. doi: 10.3390/s21165571. Sensors (Basel). 2021. PMID: 34451013 Free PMC article.
-
Hybrid Feature-Learning-Based PSO-PCA Feature Engineering Approach for Blood Cancer Classification.Diagnostics (Basel). 2023 Aug 14;13(16):2672. doi: 10.3390/diagnostics13162672. Diagnostics (Basel). 2023. PMID: 37627931 Free PMC article.
-
Optimal features selection in the high dimensional data based on robust technique: Application to different health database.Heliyon. 2024 Sep 2;10(17):e37241. doi: 10.1016/j.heliyon.2024.e37241. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39296019 Free PMC article. Review.
-
A hybrid machine learning feature selection model-HMLFSM to enhance gene classification applied to multiple colon cancers dataset.PLoS One. 2023 Nov 2;18(11):e0286791. doi: 10.1371/journal.pone.0286791. eCollection 2023. PLoS One. 2023. PMID: 37917732 Free PMC article. Review.
Cited by
-
Gene selection based on adaptive neighborhood-preserving multi-objective particle swarm optimization.PeerJ Comput Sci. 2025 May 28;11:e2872. doi: 10.7717/peerj-cs.2872. eCollection 2025. PeerJ Comput Sci. 2025. PMID: 40567808 Free PMC article.
References
-
- S. Vanjimalar, D. Ramyachitra, P. Manikandan. A review on feature selection techniques for gene expression data. In: 2018 IEEE International Conference on Computational Intelligence and Computing Research (ICCIC). 2018. p. 1–4.
-
- Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics (Oxford, England) 2007;23:2507–2517. - PubMed
-
- George V, Velanganny C. Review on feature selection techniques and the impact of Svm for cancer classification using gene expression profile. Int J Comput Sci Eng Surv. 2011;2:16–27.
-
- Zhu Z, Ong Y-S, Dash M. Markov blanket-embedded genetic algorithm for gene selection. Pattern Recognit. 2007;40(11):3236–3248.
-
- Li S, Wu X, Hu X. Gene selection using genetic algorithm and support vectors machines. Soft Comput. 2008;12(7):693–698.
MeSH terms
LinkOut - more resources
Full Text Sources

