A two-stage genome-wide association study of radiation-induced acute toxicity in head and neck cancer
- PMID: 34838041
- PMCID: PMC8626989
- DOI: 10.1186/s12967-021-03145-1
A two-stage genome-wide association study of radiation-induced acute toxicity in head and neck cancer
Abstract
Background: Most head and neck cancer (HNC) patients receive radiotherapy (RT) and develop toxicities. This genome-wide association study (GWAS) was designed to identify single nucleotide polymorphisms (SNPs) associated with common acute radiation-induced toxicities (RITs) in an HNC cohort.
Methods: A two-stage GWAS was performed in 1279 HNC patients treated with RT and prospectively scored for mucositis, xerostomia, sticky saliva, and dysphagia. The area under the curve (AUC) was used to estimate the average load of toxicity during RT. At the discovery study, multivariate linear regression was used in 957 patients, and the top-ranking SNPs were tested in 322 independent replication cohort. Next, the discovery and the replication studies were meta-analyzed.
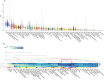
Results: A region on 5q21.3 containing 16 SNPs showed genome-wide (GW) significance association at P-value < 5.0 × 10-8 with patient-rated acute xerostomia in the discovery study. The top signal was rs35542 with an adjusted effect size of 0.17*A (95% CI 0.12 to 0.23; P-value < = 3.78 × 10-9). The genome wide significant SNPs were located within three genes (EFNA5, FBXL17, and FER). In-silico functional analysis showed these genes may be involved in DNA damage response and co-expressed in minor salivary glands. We found 428 suggestive SNPs (P-value < 1.0 × 10-5) for other toxicities, taken to the replication study. Eleven of them showed a nominal association (P-value < 0.05).
Conclusions: This GWAS suggested novel SNPs for patient-rated acute xerostomia in HNC patients. If validated, these SNPs and their related functional pathways could lead to a predictive assay to identify sensitive patients to radiation, which may eventually allow a more individualized RT treatment.
Keywords: GWAS; Head and neck cancer; Radiation-induced toxicity; Radiogenomics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Brodin NP, Kabarriti R, Garg MK, Guha C, Tomé WA. Systematic review of normal tissue complication models relevant to standard fractionation radiation therapy of the head and neck region published after the QUANTEC reports. Int J Radiat Oncol Biol Phys. 2018;100(2):391–407. doi: 10.1016/j.ijrobp.2017.09.041. - DOI - PMC - PubMed
-
- Sroussi HY, Epstein JB, Bensadoun RJ, Saunders DP, Lalla RV, Migliorati CA, et al. Common oral complications of head and neck cancer radiation therapy: mucositis, infections, saliva change, fibrosis, sensory dysfunctions, dental caries, periodontal disease, and osteoradionecrosis. Cancer Med. 2017;6(12):2918–2931. doi: 10.1002/cam4.1221. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

