A panel of CSF proteins separates genetic frontotemporal dementia from presymptomatic mutation carriers: a GENFI study
- PMID: 34838088
- PMCID: PMC8626910
- DOI: 10.1186/s13024-021-00499-4
A panel of CSF proteins separates genetic frontotemporal dementia from presymptomatic mutation carriers: a GENFI study
Abstract
Background: A detailed understanding of the pathological processes involved in genetic frontotemporal dementia is critical in order to provide the patients with an optimal future treatment. Protein levels in CSF have the potential to reflect different pathophysiological processes in the brain. We aimed to identify and evaluate panels of CSF proteins with potential to separate symptomatic individuals from individuals without clinical symptoms (unaffected), as well as presymptomatic individuals from mutation non-carriers.
Methods: A multiplexed antibody-based suspension bead array was used to analyse levels of 111 proteins in CSF samples from 221 individuals from families with genetic frontotemporal dementia. The data was explored using LASSO and Random forest.
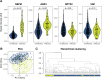
Results: When comparing affected individuals with unaffected individuals, 14 proteins were identified as potentially important for the separation. Among these, four were identified as most important, namely neurofilament medium polypeptide (NEFM), neuronal pentraxin 2 (NPTX2), neurosecretory protein VGF (VGF) and aquaporin 4 (AQP4). The combined profile of these four proteins successfully separated the two groups, with higher levels of NEFM and AQP4 and lower levels of NPTX2 in affected compared to unaffected individuals. VGF contributed to the models, but the levels were not significantly lower in affected individuals. Next, when comparing presymptomatic GRN and C9orf72 mutation carriers in proximity to symptom onset with mutation non-carriers, six proteins were identified with a potential to contribute to a separation, including progranulin (GRN).
Conclusion: In conclusion, we have identified several proteins with the combined potential to separate affected individuals from unaffected individuals, as well as proteins with potential to contribute to the separation between presymptomatic individuals and mutation non-carriers. Further studies are needed to continue the investigation of these proteins and their potential association to the pathophysiological mechanisms in genetic FTD.
Keywords: Aquaporin 4 (AQP4); Cerebrospinal fluid; LASSO; Neurofilament medium polypeptide (NEFM); Neuronal pentraxin 2 (NPTX2); Neurosecretory protein VGF (VGF); Random forest; Suspension bead array.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interest relevant to the work conducted in the presented study.
Figures

References
-
- Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Holtzman DM, Jagust W, Jessen F, Karlawish J, Liu E, Molinuevo JL, Montine T, Phelps C, Rankin KP, Rowe CC, Scheltens P, Siemers E, Snyder HM, Sperling R, Contributors. Elliott C, Masliah E, Ryan L, Silverberg N. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14(4):535–562. doi: 10.1016/j.jalz.2018.02.018. - DOI - PMC - PubMed
-
- Swift IJ, Sogorb-Esteve A, Heller C, Synofzik M, Otto M, Graff C, Galimberti D, Todd E, Heslegrave AJ, van der Ende EL, van Swieten JC, Zetterberg H, Rohrer JD. Fluid biomarkers in frontotemporal dementia: past, present and future. J Neurol Neurosurg Psychiatry. 2021;92(2):204–215. doi: 10.1136/jnnp-2020-323520. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

