The Aliment to Bodily Condition knowledgebase (ABCkb): a database connecting plants and human health
- PMID: 34838100
- PMCID: PMC8627056
- DOI: 10.1186/s13104-021-05835-x
The Aliment to Bodily Condition knowledgebase (ABCkb): a database connecting plants and human health
Abstract
Objective: Overconsumption of processed foods has led to an increase in chronic diet-related diseases such obesity and type 2 diabetes. Although diets high in fresh fruits and vegetables are linked with healthier outcomes, the specific mechanisms for these relationships are poorly understood. Experiments examining plant phytochemical production and breeding programs, or separately on the health effects of nutritional supplements have yielded results that are sparse, siloed, and difficult to integrate between the domains of human health and agriculture. To connect plant products to health outcomes through their molecular mechanism an integrated computational resource is necessary.
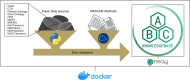
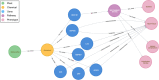
Results: We created the Aliment to Bodily Condition Knowledgebase (ABCkb) to connect plants to human health by creating a stepwise path from plant [Formula: see text] plant product [Formula: see text] human gene [Formula: see text] pathways [Formula: see text] indication. ABCkb integrates 11 curated sources as well as relationships mined from Medline abstracts by loading into a graph database which is deployed via a Docker container. This new resource, provided in a queryable container with a user-friendly interface connects plant products with human health outcomes for generating nutritive hypotheses. All scripts used are available on github ( https://github.com/atrautm1/ABCkb ) along with basic directions for building the knowledgebase and a browsable interface is available ( https://abckb.charlotte.edu ).
Keywords: Graph database; Knowledge discovery; Knowledgebase; Neo4j; Precision nutrition; Text mining.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
IBDDB: a manually curated and text-mining-enhanced database of genes involved in inflammatory bowel disease.Database (Oxford). 2021 Apr 30;2021:baab022. doi: 10.1093/database/baab022. Database (Oxford). 2021. PMID: 33929018 Free PMC article.
-
Structured reviews for data and knowledge-driven research.Database (Oxford). 2020 Jan 1;2020:baaa015. doi: 10.1093/database/baaa015. Database (Oxford). 2020. PMID: 32283553 Free PMC article.
-
T2D@ZJU: a knowledgebase integrating heterogeneous connections associated with type 2 diabetes mellitus.Database (Oxford). 2013 Jul 11;2013:bat052. doi: 10.1093/database/bat052. Print 2013. Database (Oxford). 2013. PMID: 23846596 Free PMC article.
-
Our landscapes, our livestock, ourselves: Restoring broken linkages among plants, herbivores, and humans with diets that nourish and satiate.Appetite. 2015 Dec;95:500-19. doi: 10.1016/j.appet.2015.08.004. Epub 2015 Aug 4. Appetite. 2015. PMID: 26247703 Review.
-
Plant Reactome and PubChem: The Plant Pathway and (Bio)Chemical Entity Knowledgebases.Methods Mol Biol. 2022;2443:511-525. doi: 10.1007/978-1-0716-2067-0_27. Methods Mol Biol. 2022. PMID: 35037224 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

