Whole-genome analysis reveals the contribution of non-coding de novo transposon insertions to autism spectrum disorder
- PMID: 34838103
- PMCID: PMC8627061
- DOI: 10.1186/s13100-021-00256-w
Whole-genome analysis reveals the contribution of non-coding de novo transposon insertions to autism spectrum disorder
Abstract
Background: Retrotransposons have been implicated as causes of Mendelian disease, but their role in autism spectrum disorder (ASD) has not been systematically defined, because they are only called with adequate sensitivity from whole genome sequencing (WGS) data and a large enough cohort for this analysis has only recently become available.
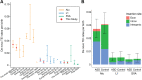
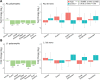
Results: We analyzed WGS data from a cohort of 2288 ASD families from the Simons Simplex Collection by establishing a scalable computational pipeline for retrotransposon insertion detection. We report 86,154 polymorphic retrotransposon insertions-including > 60% not previously reported-and 158 de novo retrotransposition events. The overall burden of de novo events was similar between ASD individuals and unaffected siblings, with 1 de novo insertion per 29, 117, and 206 births for Alu, L1, and SVA respectively, and 1 de novo insertion per 21 births total. However, ASD cases showed more de novo L1 insertions than expected in ASD genes. Additionally, we observed exonic insertions in loss-of-function intolerant genes, including a likely pathogenic exonic insertion in CSDE1, only in ASD individuals.
Conclusions: These findings suggest a modest, but important, impact of intronic and exonic retrotransposon insertions in ASD, show the importance of WGS for their analysis, and highlight the utility of specific bioinformatic tools for high-throughput detection of retrotransposon insertions.
Keywords: Alu; Autism spectrum disorder; LINE-1; Neurobiology; Polymorphic insertions; Retrotransposons; SVA; Transposable elements; de novo insertions; de novo rates.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Identification of polymorphic SVA retrotransposons using a mobile element scanning method for SVA (ME-Scan-SVA).Mob DNA. 2016 Jul 30;7:15. doi: 10.1186/s13100-016-0072-x. eCollection 2016. Mob DNA. 2016. PMID: 27478512 Free PMC article.
-
RDA coupled with deep sequencing detects somatic SVA-retrotranspositions and mosaicism in the human brain.Front Cell Dev Biol. 2023 Jun 1;11:1201258. doi: 10.3389/fcell.2023.1201258. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37325565 Free PMC article.
-
Whole Exome Sequencing Identifies Novel De Novo Variants Interacting with Six Gene Networks in Autism Spectrum Disorder.Genes (Basel). 2020 Dec 22;12(1):1. doi: 10.3390/genes12010001. Genes (Basel). 2020. PMID: 33374967 Free PMC article.
-
Roles for retrotransposon insertions in human disease.Mob DNA. 2016 May 6;7:9. doi: 10.1186/s13100-016-0065-9. eCollection 2016. Mob DNA. 2016. PMID: 27158268 Free PMC article. Review.
-
Functional impact of the human mobilome.Curr Opin Genet Dev. 2013 Jun;23(3):264-70. doi: 10.1016/j.gde.2013.02.007. Epub 2013 Mar 22. Curr Opin Genet Dev. 2013. PMID: 23523050 Free PMC article. Review.
Cited by
-
Intragenic L1 Insertion: One Possibility of Brain Disorder.Life (Basel). 2022 Sep 13;12(9):1425. doi: 10.3390/life12091425. Life (Basel). 2022. PMID: 36143463 Free PMC article. Review.
-
Integrating Genetic Structural Variations and Whole-Genome Sequencing Into Clinical Neurology.Neurol Genet. 2022 May 27;8(4):e200005. doi: 10.1212/NXG.0000000000200005. eCollection 2022 Aug. Neurol Genet. 2022. PMID: 37435434 Free PMC article. Review.
-
C1q/TNF-related protein-1: Potential biomarker for early diagnosis of autism spectrum disorder.Int J Immunopathol Pharmacol. 2022 Jan-Dec;36:3946320221079471. doi: 10.1177/03946320221079471. Int J Immunopathol Pharmacol. 2022. PMID: 35202556 Free PMC article.
-
Impaired OTUD7A-dependent Ankyrin regulation mediates neuronal dysfunction in mouse and human models of the 15q13.3 microdeletion syndrome.Mol Psychiatry. 2023 Apr;28(4):1747-1769. doi: 10.1038/s41380-022-01937-5. Epub 2023 Jan 6. Mol Psychiatry. 2023. PMID: 36604605 Free PMC article.
-
Transposable elements are dynamically regulated in medium spiny neurons and may contribute to the molecular and behavioral adaptations to cocaine.Biol Psychiatry. 2025 Jul 25:S0006-3223(25)01352-6. doi: 10.1016/j.biopsych.2025.07.014. Online ahead of print. Biol Psychiatry. 2025. PMID: 40716507
References
-
- Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373(Supplement C):134–137. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

