3D hemisphere-based convolutional neural network for whole-brain MRI segmentation
- PMID: 34839147
- PMCID: PMC10116838
- DOI: 10.1016/j.compmedimag.2021.102000
3D hemisphere-based convolutional neural network for whole-brain MRI segmentation
Abstract
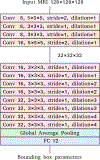
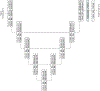
Whole-brain segmentation is a crucial pre-processing step for many neuroimaging analyses pipelines. Accurate and efficient whole-brain segmentations are important for many neuroimage analysis tasks to provide clinically relevant information. Several recently proposed convolutional neural networks (CNN) perform whole brain segmentation using individual 2D slices or 3D patches as inputs due to graphical processing unit (GPU) memory limitations, and use sliding windows to perform whole brain segmentation during inference. However, these approaches lack global and spatial information about the entire brain and lead to compromised efficiency during both training and testing. We introduce a 3D hemisphere-based CNN for automatic whole-brain segmentation of T1-weighted magnetic resonance images of adult brains. First, we trained a localization network to predict bounding boxes for both hemispheres. Then, we trained a segmentation network to segment one hemisphere, and segment the opposing hemisphere by reflecting it across the mid-sagittal plane. Our network shows high performance both in terms of segmentation efficiency and accuracy (0.84 overall Dice similarity and 6.1 mm overall Hausdorff distance) in segmenting 102 brain structures. On multiple independent test datasets, our method demonstrated a competitive performance in the subcortical segmentation task and a high consistency in volumetric measurements of intra-session scans.
Keywords: 3D CNN; MRI; Segmentation.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Figures




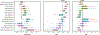




References
-
- de Brebisson A, Montana G, 2015. Deep neural networks for anatomical brain segmentation, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 20–28.
-
- Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL, 2014. Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv preprint arXiv:1412.7062. - PubMed
-
- Derakhshan M, Caramanos Z, Giacomini PS, Narayanan S, Maranzano J, Francis SJ, Arnold DL, Collins DL, 2010. Evaluation of automated techniques for the quantification of grey matter atrophy in patients with multiple sclerosis. NeuroImage 52, 1261–1267. doi:10.1016/j.neuroimage.2010.05.029. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

