Identification of miRNA Regulatory Networks and Candidate Markers for Fracture Healing in Mice
- PMID: 34840596
- PMCID: PMC8611357
- DOI: 10.1155/2021/2866475
Identification of miRNA Regulatory Networks and Candidate Markers for Fracture Healing in Mice
Abstract
Background: It is important to improve the understanding of the fracture healing process at the molecular levels, then to discover potential miRNA regulatory mechanisms and candidate markers.
Methods: Expression profiles of mRNA and miRNA were obtained from the Gene Expression Omnibus database. We performed differential analysis, enrichment analysis, protein-protein interaction (PPI) network analysis. The miRNA-mRNA network analysis was also performed.
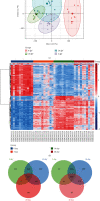
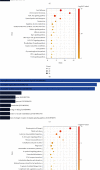
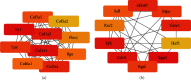
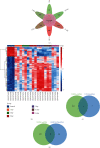
Results: We identified 499 differentially expressed mRNAs (DEmRs) that were upregulated and 534 downregulated DEmRs during fracture healing. They were mainly enriched in collagen fibril organization and immune response. Using the PPI network, we screened 10 hub genes that were upregulated and 10 hub genes downregulated with the largest connectivity. We further constructed the miRNA regulatory network for hub genes and identified 13 differentially expressed miRNAs (DEmiRs) regulators. Cd19 and Col6a1 were identified as key candidate mRNAs with the largest fold change, and their DEmiR regulators were key candidate regulators.
Conclusion: Cd19 and Col6a1 might serve as candidate markers for fracture healing in subsequent studies. Their expression is regulated by miRNAs and is involved in collagen fibril organization and immune responses.
Copyright © 2021 Xianglu Li et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

