Proteome Analysis of the Hypothalamic Arcuate Nucleus in Chronic High-Fat Diet-Induced Obesity
- PMID: 34840970
- PMCID: PMC8617565
- DOI: 10.1155/2021/3501770
Proteome Analysis of the Hypothalamic Arcuate Nucleus in Chronic High-Fat Diet-Induced Obesity
Abstract
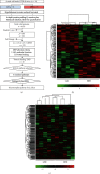
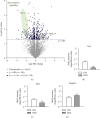
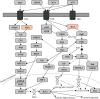
The hypothalamus plays a central role in the integrated regulation of feeding and energy homeostasis. The hypothalamic arcuate nucleus (ARC) contains a population of neurons that express orexigenic and anorexigenic factors and is thought to control feeding behavior via several neuronal circuits. In this study, a comparative proteomic analysis of low-fat control diet- (LFD-) and high-fat diet- (HFD-) induced hypothalamic ARC was performed to identify differentially expressed proteins (DEPs) related to changes in body weight. In the ARC in the hypothalamus, 6621 proteins (FDR < 0.01) were detected, and 178 proteins were categorized as DEPs (89 upregulated and 89 downregulated in the HFD group). Among the Gene Ontology molecular function terms associated with the DEPs, protein binding was the most significant. Fibroblast growth factor receptor substrate 2 (Frs2) and SHC adaptor protein 3 (Shc3) were related to protein binding and involved in the neurotrophin signaling pathway according to Kyoto Encyclopedia of Genes and Genomes analysis. Furthermore, high-precision quantitative proteomic analysis revealed that the protein profile of the ARC in mice with HFD-induced obesity differed from that in LFD mice, thereby offering insight into the molecular basis of feeding regulation and suggesting Frs2 and Shc3 as novel treatment targets for central anorexigenic signal induction.
Copyright © 2021 Chang Yeon Kim et al.
Conflict of interest statement
The authors declare no conflicts of interest regarding the publication of this paper.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

