Various short autonomously replicating sequences from the yeast Kluyveromyces marxianus seemingly without canonical consensus
- PMID: 34841344
- PMCID: PMC8610295
- DOI: 10.1016/j.crmicr.2021.100053
Various short autonomously replicating sequences from the yeast Kluyveromyces marxianus seemingly without canonical consensus
Abstract
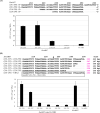
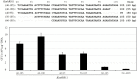
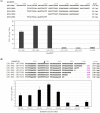
Eukaryotic autonomously replicating sequences (ARSs) are composed of three domains, A, B, and C. Domain A is comprised of an ARS consensus sequence (ACS), while the B domain has the DNA unwinding element and the C domain is important for DNA-protein interactions. In Saccharomyces cerevisiae and Kluyveromyces lactis ARS101, the ACS is commonly composed of 11 bp, 5'-(A/T)AAA(C/T)ATAAA(A/T)-3'. This core sequence is essential for S. cerevisiae and K. lactis ARS activity. In this study, we identified ARS-containing sequences from genomic libraries of the yeast Kluyveromyces marxianus DMKU3-1042 and validated their replication activities. The identified K. marxianus DMKU3-1042 ARSs (KmARSs) have very effective replication ability but their sequences are divergent and share no common consensus. We have carried out point mutations, deletions, and base pairs substitutions within the sequences of some of the KmARSs to identify the sequence(s) that influence the replication activity. Consensus sequences same as the 11 bp ACS of S. cerevisiae and K. lactis were not found in all minimum functional KmARSs reported here except KmARS7. Moreover, partial sequences from different KmARSs are interchangeable among each other to retain the ARS activity. We have also specifically identified the essential nucleotides, which are indispensable for replication, within some of the KmARSs. Our deletions analysis revealed that only 21 bp in KmARS18 could retain the ARS activity. The identified KmARSs in this study are unique compared to other yeasts' ARSs, do not share common ACS, and are interchangeable.
Keywords: Functional validation; Interchanged sequences; KmARS; NHEJ; Transformability.
© 2021 The Authors. Published by Elsevier B.V.
Conflict of interest statement
None
Figures



References
-
- Abdel-Banat B.M.A., Hoshida H., Ano A., Nonklang S., Akada R. High-temperature fermentation: how can processes for ethanol production at high temperatures become superior to the traditional process using mesophilic yeast? Appl. Microbiol. Biotechnol. 2010;85:861–867. - PubMed
-
- Abdel-Banat B.M.A., Nonklang S., Hoshida H., Akada R. Random and targeted gene integrations through the control of non-homologous end joining in the yeast Kluyveromyces marxianus. Yeast. 2010;27:29–39. - PubMed
-
- Akada R., Kitagawa T., Kaneko S., Toyonaga D., Ito S., Kakihara Y, Hoshida H., Morimura S., Kondo A, Kida K. PCR-mediated seamless gene deletion and marker recycling in Saccharomyces cerevisiae. Yeast. 2006;23:399–405. - PubMed
-
- Ausubel F.M, Brent R., Kingston R.E., Moore D.D. Wiley; New York: 1999. Short Protocols in Molecular Biology.
-
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
LinkOut - more resources
Full Text Sources

