Changes in glucagon-like peptide 1 and 2 levels in people with obesity after a diet-induced weight-loss intervention are related to a specific microbiota signature: A prospective cohort study
- PMID: 34841718
- PMCID: PMC8571947
- DOI: 10.1002/ctm2.575
Changes in glucagon-like peptide 1 and 2 levels in people with obesity after a diet-induced weight-loss intervention are related to a specific microbiota signature: A prospective cohort study
Conflict of interest statement
The authors declare no conflict of interest.
Figures

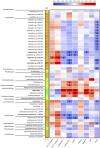
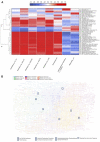
References
Publication types
MeSH terms
Substances
Grants and funding
- PI14/00228/Instituto de Salud Carlos III
- PI17/0153/Instituto de Salud Carlos III
- PI20/00338/Instituto de Salud Carlos III
- RTI2018-093919-B-I00/Ministerio de Ciencia e Innovación
- PID2019-105969GB-I00/Ministerio de Ciencia e Innovación
- CB07708/0012/Spanish Biomedical Research Center in Diabetes and Associated Metabolic Disorders
- Instituto de Salud Carlos III. S.F-V./Spanish Biomedical Research Center in Diabetes and Associated Metabolic Disorders
- CP10/00438/Spanish Biomedical Research Center in Diabetes and Associated Metabolic Disorders
- CPII16/00008/Spanish Biomedical Research Center in Diabetes and Associated Metabolic Disorders
- PI14/00228/Instituto de Salud Carlos III
- PI17/0153/Instituto de Salud Carlos III
- RTI2018-093919-B-I00/Ministerio de Ciencia e Innovación
- PID2019-105969GB-I00/Ministerio de Ciencia e Innovación
- PROMETEO/2018/133/Generalitat Valenciana
- CB07708/0012/Centro de Investigación Biomédica en Red Diabetes y Enfermedades Metabólicas Asociadas
LinkOut - more resources
Full Text Sources
Medical
