Analysis of changes in microbiome compositions related to the prognosis of colorectal cancer patients based on tissue-derived 16S rRNA sequences
- PMID: 34844611
- PMCID: PMC8628381
- DOI: 10.1186/s12967-021-03154-0
Analysis of changes in microbiome compositions related to the prognosis of colorectal cancer patients based on tissue-derived 16S rRNA sequences
Abstract
Background: Comparing the microbiome compositions obtained under different physiological conditions has frequently been attempted in recent years to understand the functional influence of microbiomes in the occurrence of various human diseases.
Methods: In the present work, we analyzed 102 microbiome datasets containing tumor- and normal tissue-derived microbiomes obtained from a total of 51 Korean colorectal cancer (CRC) patients using 16S rRNA amplicon sequencing. Two types of comparisons were used: 'normal versus (vs.) tumor' comparison and 'recurrent vs. nonrecurrent' comparison, for which the prognosis of patients was retrospectively determined.
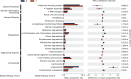
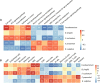
Results: As a result, we observed that in the 'normal vs. tumor' comparison, three phyla, Firmicutes, Actinobacteria, and Bacteroidetes, were more abundant in normal tissues, whereas some pathogenic bacteria, including Fusobacterium nucleatum and Bacteroides fragilis, were more abundant in tumor tissues. We also found that bacteria with metabolic pathways related to the production of bacterial motility proteins or bile acid secretion were more enriched in tumor tissues. In addition, the amount of these two pathogenic bacteria was positively correlated with the expression levels of host genes involved in the cell cycle and cell proliferation, confirming the association of microbiomes with tumorigenic pathway genes in the host. Surprisingly, in the 'recurrent vs. nonrecurrent' comparison, we observed that these two pathogenic bacteria were more abundant in the patients without recurrence than in the patients with recurrence. The same conclusion was drawn in the analysis of both normal and tumor-derived microbiomes.
Conclusions: Taken together, it seems that understanding the composition of tissue microbiomes is useful for predicting the prognosis of CRC patients.
Keywords: 16S rRNA; B. fragilis; Colorectal cancer; F. nucleatum; Microbiome.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Macrae FA. Colorectal cancer: epidemiology, risk factors, and protective factors. Uptodate com [ažurirano 9.lipnja 2017]. 2016.
-
- Sandler RS. Epidemiology and risk factors for colorectal cancer. Gastroenterol Clin N Am. 1996;25(4):717–735. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

