Temporal and sequential order of nonoverlapping gene networks unraveled in mated female Drosophila
- PMID: 34844981
- PMCID: PMC8645335
- DOI: 10.26508/lsa.202101119
Temporal and sequential order of nonoverlapping gene networks unraveled in mated female Drosophila
Abstract
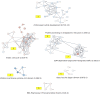
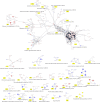
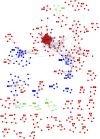
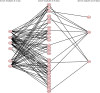
In this study, we reanalyzed available datasets of gene expression changes in female Drosophila head induced by mating. Mated females present metabolic phenotypic changes and display behavioral characteristics that are not observed in virgin females, such as repulsion to male sexual aggressiveness, fidelity to food spots selected for oviposition, and restriction to the colonization of new niches. We characterize gene networks that play a role in female brain plasticity after mating using AMINE, a novel algorithm to find dysregulated modules of interacting genes. The uncovered networks of altered genes revealed a strong specificity for each successive period of life span after mating in the female head, with little conservation between them. This finding highlights a temporal order of recruitment of waves of interconnected genes which are apparently transiently modified: the first wave disappears before the emergence of the second wave in a reversible manner and ends with few consolidated gene expression changes at day 20. This analysis might document an extended field of a programmatic control of female phenotypic traits by male seminal fluid.
© 2021 Pasquier et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
