Metagenomic discovery of CRISPR-associated transposons
- PMID: 34845024
- PMCID: PMC8670466
- DOI: 10.1073/pnas.2112279118
Metagenomic discovery of CRISPR-associated transposons
Abstract
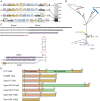
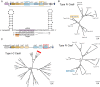
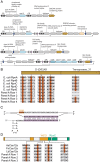
CRISPR-associated Tn7 transposons (CASTs) co-opt cas genes for RNA-guided transposition. CASTs are exceedingly rare in genomic databases; recent surveys have reported Tn7-like transposons that co-opt Type I-F, I-B, and V-K CRISPR effectors. Here, we expand the diversity of reported CAST systems via a bioinformatic search of metagenomic databases. We discover architectures for all known CASTs, including arrangements of the Cascade effectors, target homing modalities, and minimal V-K systems. We also describe families of CASTs that have co-opted the Type I-C and Type IV CRISPR-Cas systems. Our search for non-Tn7 CASTs identifies putative candidates that include a nuclease dead Cas12. These systems shed light on how CRISPR systems have coevolved with transposases and expand the programmable gene-editing toolkit.
Keywords: CAST; CRISPR RNA; bioinformatics; gene editing; transposition.
Conflict of interest statement
Competing interest statement: The authors are coinventors on patent applications filed based on this work.
Figures



References
-
- Halpin-Healy T. S., Klompe S. E., Sternberg S. H., Fernández I. S., Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system. Nature 577, 271–274 (2020). - PubMed
-
- Klompe S. E., Vo P. L. H., Halpin-Healy T. S., Sternberg S. H., Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

