Multi-species host range of staphylococcal phages isolated from wastewater
- PMID: 34845206
- PMCID: PMC8629997
- DOI: 10.1038/s41467-021-27037-6
Multi-species host range of staphylococcal phages isolated from wastewater
Abstract
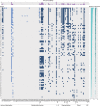
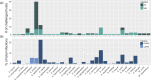
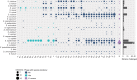
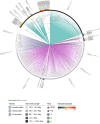
The host range of bacteriophages defines their impact on bacterial communities and genome diversity. Here, we characterize 94 novel staphylococcal phages from wastewater and establish their host range on a diversified panel of 117 staphylococci from 29 species. Using this high-resolution phage-bacteria interaction matrix, we unveil a multi-species host range as a dominant trait of the isolated staphylococcal phages. Phage genome sequencing shows this pattern to prevail irrespective of taxonomy. Network analysis between phage-infected bacteria reveals that hosts from multiple species, ecosystems, and drug-resistance phenotypes share numerous phages. Lastly, we show that phages throughout this network can package foreign genetic material enclosing an antibiotic resistance marker at various frequencies. Our findings indicate a weak host specialism of the tested phages, and therefore their potential to promote horizontal gene transfer in this environment.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bernheim A, Sorek R. The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol. 2020;18:113–119. - PubMed
-
- de Jonge PA, Nobrega FL, Brouns SJJ, Dutilh BE. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2019;27:51–63. - PubMed
-
- Dion MB, Oechslin F, Moineau S. Phage diversity, genomics, and phylogeny. Nat. Rev. Microbiol. 2020;18:125–138. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

