Representation and participation across 20 years of plant genome sequencing
- PMID: 34845350
- PMCID: PMC8677620
- DOI: 10.1038/s41477-021-01031-8
Representation and participation across 20 years of plant genome sequencing
Abstract
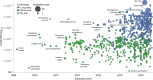
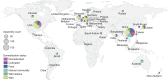
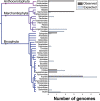
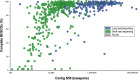
The field of plant genome sequencing has grown rapidly in the past 20 years, leading to increases in the quantity and quality of publicly available genomic resources. The growing wealth of genomic data from an increasingly diverse set of taxa provides unprecedented potential to better understand the genome biology and evolution of land plants. Here we provide a contemporary view of land plant genomics, including analyses on assembly quality, taxonomic distribution of sequenced species and national participation. We show that assembly quality has increased dramatically in recent years, that substantial taxonomic gaps exist and that the field has been dominated by affluent nations in the Global North and China, despite a wide geographic distribution of study species. We identify numerous disconnects between the native range of focal species and the national affiliation of the researchers studying them, which we argue are rooted in colonialism-both past and present. Luckily, falling sequencing costs, widening availability of analytical tools and an increasingly connected scientific community provide key opportunities to improve existing assemblies, fill sampling gaps and empower a more global plant genomics community.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Decolonizing botanical genomics.Nat Plants. 2021 Dec;7(12):1542-1543. doi: 10.1038/s41477-021-01041-6. Nat Plants. 2021. PMID: 34862483 No abstract available.
-
The landscape of genome sequencing and assembling in plants.Funct Integr Genomics. 2022 Dec;22(6):1147-1152. doi: 10.1007/s10142-022-00916-x. Epub 2022 Nov 15. Funct Integr Genomics. 2022. PMID: 36376541 No abstract available.
References
-
- Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Michael TP, VanBuren R. Building near-complete plant genomes. Curr. Opin. Plant Biol. 2020;54:26–33. - PubMed
-
- Sharma, P. et al. Improvements in the sequencing and assembly of plant genomes. https://gigabytejournal.com/articles/24 (2021). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

