OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies
- PMID: 34845388
- PMCID: PMC8648559
- DOI: 10.1038/s41592-021-01326-w
OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies
Abstract
The rapid pace of innovation in biological imaging and the diversity of its applications have prevented the establishment of a community-agreed standardized data format. We propose that complementing established open formats such as OME-TIFF and HDF5 with a next-generation file format such as Zarr will satisfy the majority of use cases in bioimaging. Critically, a common metadata format used in all these vessels can deliver truly findable, accessible, interoperable and reusable bioimaging data.
© 2021. The Author(s).
Conflict of interest statement
C.A., E.D., K.K., M.L. and J.R.S. are affiliated with Glencoe Software, a commercial company that builds, delivers, supports and integrates image data management systems across academic, biotech and pharmaceutical industries. The remaining authors declare no competing interests.
Figures



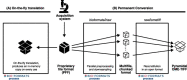
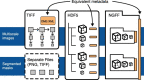
References
-
- The HDF5 Library and File Format (The HDF Group, accessed 18 October 2021); https://www.hdfgroup.org/solutions/hdf5/
-
- Miles, A. et al. zarr-developers/zarr-python: v.2.5.0 10.5281/zenodo.4069231 (2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

