Second-Shell Amino Acid R266 Helps Determine N-Succinylamino Acid Racemase Reaction Specificity in Promiscuous N-Succinylamino Acid Racemase/ o-Succinylbenzoate Synthase Enzymes
- PMID: 34845903
- PMCID: PMC8939854
- DOI: 10.1021/acs.biochem.1c00627
Second-Shell Amino Acid R266 Helps Determine N-Succinylamino Acid Racemase Reaction Specificity in Promiscuous N-Succinylamino Acid Racemase/ o-Succinylbenzoate Synthase Enzymes
Abstract
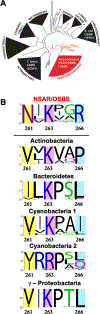
Catalytic promiscuity is the coincidental ability to catalyze nonbiological reactions in the same active site as the native biological reaction. Several lines of evidence show that catalytic promiscuity plays a role in the evolution of new enzyme functions. Thus, studying catalytic promiscuity can help identify structural features that predispose an enzyme to evolve new functions. This study identifies a potentially preadaptive residue in a promiscuous N-succinylamino acid racemase/o-succinylbenzoate synthase (NSAR/OSBS) enzyme from Amycolatopsis sp. T-1-60. This enzyme belongs to a branch of the OSBS family which includes many catalytically promiscuous NSAR/OSBS enzymes. R266 is conserved in all members of the NSAR/OSBS subfamily. However, the homologous position is usually hydrophobic in other OSBS subfamilies, whose enzymes lack NSAR activity. The second-shell amino acid R266 is close to the catalytic acid/base K263, but it does not contact the substrate, suggesting that R266 could affect the catalytic mechanism. Mutating R266 to glutamine in Amycolatopsis NSAR/OSBS profoundly reduces NSAR activity but moderately reduces OSBS activity. This is due to a 1000-fold decrease in the rate of proton exchange between the substrate and the general acid/base catalyst K263. This mutation is less deleterious for the OSBS reaction because K263 forms a cation-π interaction with the OSBS substrate and/or the intermediate, rather than acting as a general acid/base catalyst. Together, the data explain how R266 contributes to NSAR reaction specificity and was likely an essential preadaptation for the evolution of NSAR activity.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


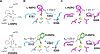

References
-
- Brodkin HR, Novak WR, Milne AC, D’Aquino JA, Karabacak NM, Goldberg IG, Agar JN, Payne MS, Petsko GA, Ondrechen MJ, and Ringe D (2011) Evidence of the participation of remote residues in the catalytic activity of Co-type nitrile hydratase from Pseudomonas putida, Biochemistry 50, 4923–4935. - PMC - PubMed
-
- Blaisse MR, Fu B, and Chang MCY (2018) Structural and Biochemical Studies of Substrate Selectivity in Ascaris suum Thiolases, Biochemistry 57, 3155–3166. - PubMed
-
- Yang G, Hong N, Baier F, Jackson CJ, and Tokuriki N (2016) Conformational Tinkering Drives Evolution of a Promiscuous Activity through Indirect Mutational Effects, Biochemistry 55, 4583–4593. - PubMed
-
- Graf L, Craik CS, Patthy A, Roczniak S, Fletterick RJ, and Rutter WJ (1987) Selective alteration of substrate specificity by replacement of aspartic acid-189 with lysine in the binding pocket of trypsin, Biochemistry 26, 2616–2623. - PubMed
-
- Perona JJ, Hedstrom L, Rutter WJ, and Fletterick RJ (1995) Structural origins of substrate discrimination in trypsin and chymotrypsin, Biochemistry 34, 1489–1499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

