Computational Modeling of the Virucidal Inhibition Mechanism for Broad-Spectrum Antiviral Nanoparticles and HPV16 Capsid Segments
- PMID: 34845905
- PMCID: PMC8926016
- DOI: 10.1021/acs.jpcb.1c07436
Computational Modeling of the Virucidal Inhibition Mechanism for Broad-Spectrum Antiviral Nanoparticles and HPV16 Capsid Segments
Abstract
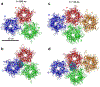
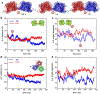
Solid core nanoparticles (NPs) coated with sulfonated ligands that mimic heparan sulfate proteoglycans (HSPGs) can exhibit virucidal activity against many viruses that utilize HSPG interactions with host cells for the initial stages of infection. How the interactions of these NPs with large capsid segments of HSPG-interacting viruses lead to their virucidal activity has been unclear. Here, we describe the interactions between sulfonated NPs and segments of the human papilloma virus type 16 (HPV16) capsids using atomistic molecular dynamics simulations. The simulations demonstrate that the NPs primarily bind at the interfaces of two HPV16 capsid proteins. After equilibration, the distances and angles between capsid proteins in the capsid segments are larger for the systems in which the NPs bind at the interfaces of capsid proteins. Over time, NP binding can lead to breaking of contacts between two neighboring proteins. The revealed mechanism of NPs targeting the interfaces between pairs of capsid proteins can be utilized for designing new generations of virucidal materials and contribute to the development of new broad-spectrum non-toxic virucidal materials.
Figures



Similar articles
-
Broad-spectrum non-toxic antiviral nanoparticles with a virucidal inhibition mechanism.Nat Mater. 2018 Feb;17(2):195-203. doi: 10.1038/nmat5053. Epub 2017 Dec 18. Nat Mater. 2018. PMID: 29251725
-
Sulfonated Nanomaterials with Broad-Spectrum Antiviral Activity Extending beyond Heparan Sulfate-Dependent Viruses.Antimicrob Agents Chemother. 2020 Nov 17;64(12):e02001-20. doi: 10.1128/AAC.02001-20. Print 2020 Nov 17. Antimicrob Agents Chemother. 2020. PMID: 32988820 Free PMC article.
-
Ligand concentration determines antiviral efficacy of silica multivalent nanoparticles.J Colloid Interface Sci. 2024 Mar;657:327-333. doi: 10.1016/j.jcis.2023.11.122. Epub 2023 Nov 23. J Colloid Interface Sci. 2024. PMID: 38043234
-
Silver nanoparticles are broad-spectrum bactericidal and virucidal compounds.J Nanobiotechnology. 2011 Aug 3;9:30. doi: 10.1186/1477-3155-9-30. J Nanobiotechnology. 2011. PMID: 21812950 Free PMC article. Review.
-
Theoretical studies of viral capsid proteins.Curr Opin Struct Biol. 2000 Apr;10(2):170-3. doi: 10.1016/s0959-440x(00)00064-6. Curr Opin Struct Biol. 2000. PMID: 10753813 Review.
Cited by
-
Sulfoglycodendron Antivirals with Scalable Architectures and Activities.J Chem Inf Model. 2024 Sep 23;64(18):7141-7151. doi: 10.1021/acs.jcim.4c00541. Epub 2024 Sep 4. J Chem Inf Model. 2024. PMID: 39230262
-
An Atomic and Molecular Insight into How PFOA Reduces α-Helicity, Compromises Substrate Binding, and Creates Binding Pockets in a Model Globular Protein.J Am Chem Soc. 2024 May 8;146(18):12766-12777. doi: 10.1021/jacs.4c02934. Epub 2024 Apr 24. J Am Chem Soc. 2024. PMID: 38656109 Free PMC article.
-
Sulfoglycodendron Antivirals with Scalable Architectures and Activities.bioRxiv [Preprint]. 2024 Aug 18:2024.08.01.606251. doi: 10.1101/2024.08.01.606251. bioRxiv. 2024. Update in: J Chem Inf Model. 2024 Sep 23;64(18):7141-7151. doi: 10.1021/acs.jcim.4c00541. PMID: 39131386 Free PMC article. Updated. Preprint.
-
Interfacial Interactions between Nanoplastics and Biological Systems: toward an Atomic and Molecular Understanding of Plastics-Driven Biological Dyshomeostasis.ACS Appl Mater Interfaces. 2024 May 22;16(20):25740-25756. doi: 10.1021/acsami.4c03008. Epub 2024 May 9. ACS Appl Mater Interfaces. 2024. PMID: 38722759 Free PMC article.
References
-
- De Clercq E & Herdewijn P, in Pharm. Sci. Encycl 1–56 (American Cancer Society, 2010). DOI: 10.1002/9780470571224.pse026 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

