TCR meta-clonotypes for biomarker discovery with tcrdist3 enabled identification of public, HLA-restricted clusters of SARS-CoV-2 TCRs
- PMID: 34845983
- PMCID: PMC8631793
- DOI: 10.7554/eLife.68605
TCR meta-clonotypes for biomarker discovery with tcrdist3 enabled identification of public, HLA-restricted clusters of SARS-CoV-2 TCRs
Abstract
T-cell receptors (TCRs) encode clinically valuable information that reflects prior antigen exposure and potential future response. However, despite advances in deep repertoire sequencing, enormous TCR diversity complicates the use of TCR clonotypes as clinical biomarkers. We propose a new framework that leverages experimentally inferred antigen-associated TCRs to form meta-clonotypes - groups of biochemically similar TCRs - that can be used to robustly quantify functionally similar TCRs in bulk repertoires across individuals. We apply the framework to TCR data from COVID-19 patients, generating 1831 public TCR meta-clonotypes from the SARS-CoV-2 antigen-associated TCRs that have strong evidence of restriction to patients with a specific human leukocyte antigen (HLA) genotype. Applied to independent cohorts, meta-clonotypes targeting these specific epitopes were more frequently detected in bulk repertoires compared to exact amino acid matches, and 59.7% (1093/1831) were more abundant among COVID-19 patients that expressed the putative restricting HLA allele (false discovery rate [FDR]<0.01), demonstrating the potential utility of meta-clonotypes as antigen-specific features for biomarker development. To enable further applications, we developed an open-source software package, tcrdist3, that implements this framework and facilitates flexible workflows for distance-based TCR repertoire analysis.
Keywords: SARS-CoV-2; T cell receptor; biomarkers; computational biology; human; immune repertoire; immunology; inflammation; software; systems biology.
© 2021, Mayer-Blackwell et al.
Conflict of interest statement
KM, SS, LC, AS, JG, AF No competing interests declared, JC JCC served as unpaid consultant for 10X Genomics on the initial analysis of the 10x_200k dataset. TH TH has equity in Poold Diagnostics. PT is on the Scientific Advisory Boards of Immunoscape and Cytoagents, consulted for Elevate Bio and PACT Pharma, and has received travel costs and speaking fees from 10X Genomics and Illumina. PT served as unpaid consultant for 10X Genomics on the initial analysis of the 10x_200k dataset. PT also has filed patents on methods for sequencing and cloning TCRs (International PCT applications published December 24, 2020 as WO 2020/257575 and January 7, 2021 as WO 2021/003114). These applications are pending and have not yet been granted. PB served as unpaid consultant for 10X Genomics on the initial analysis of the 10x_200k dataset.
Figures





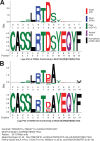


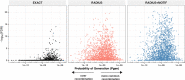
Update of
-
TCR meta-clonotypes for biomarker discovery with tcrdist3: identification of public, HLA-restricted SARS-CoV-2 associated TCR features.bioRxiv [Preprint]. 2021 Mar 19:2020.12.24.424260. doi: 10.1101/2020.12.24.424260. bioRxiv. 2021. Update in: Elife. 2021 Nov 30;10:e68605. doi: 10.7554/eLife.68605. PMID: 33398288 Free PMC article. Updated. Preprint.
References
-
- Ahmadzadeh M, Pasetto A, Jia L, Deniger DC, Stevanović S, Robbins PF, Rosenberg SA. Tumor-infiltrating human CD4+ regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Science Immunology. 2019;4:eaao4310. doi: 10.1126/sciimmunol.aao4310. - DOI - PMC - PubMed
-
- Britanova OV, Shugay M, Merzlyak EM, Staroverov DB, Putintseva EV, Turchaninova MA, Mamedov IZ, Pogorelyy MV, Bolotin DA, Izraelson M, Davydov AN, Egorov ES, Kasatskaya SA, Rebrikov DV, Lukyanov S, Chudakov DM. Dynamics of Individual T Cell Repertoires: From Cord Blood to Centenarians. Journal of Immunology. 2016;196:5005–5013. doi: 10.4049/jimmunol.1600005. - DOI - PubMed
-
- Cao K, Wu J, Li X, Xie H, Tang C, Zhao X, Wang S, Chen L, Zhang W, An Y, Li X, Lin L, Chai R, Fang M, Yue Y, Wang X, Ding Y, Zhou L, Zhao Q, Yang H, Wang J, He S, Liu X. T-cell receptor repertoire data provides new evidence for hygiene hypothesis of allergic diseases. Allergy. 2020;75:681–683. doi: 10.1111/all.14014. - DOI - PubMed
-
- Chiou S-H, Tseng D, Reuben A, Mallajosyula V, Molina IS, Conley S, Wilhelmy J, McSween AM, Yang X, Nishimiya D, Sinha R, Nabet BY, Wang C, Shrager JB, Berry MF, Backhus L, Lui NS, Wakelee HA, Neal JW, Padda SK, Berry GJ, Delaidelli A, Sorensen PH, Sotillo E, Tran P, Benson JA, Richards R, Labanieh L, Klysz DD, Louis DM, Feldman SA, Diehn M, Weissman IL, Zhang J, Wistuba II, Futreal PA, Heymach JV, Garcia KC, Mackall CL, Davis MM. Global analysis of shared T cell specificities in human non-small cell lung cancer enables HLA inference and antigen discovery. Immunity. 2021;54:586–602. doi: 10.1016/j.immuni.2021.02.014. - DOI - PMC - PubMed
-
- Coles CH, Mulvaney RM, Malla S, Walker A, Smith KJ, Lloyd A, Lowe KL, McCully ML, Martinez Hague R, Aleksic M, Harper J, Paston SJ, Donnellan Z, Chester F, Wiederhold K, Robinson RA, Knox A, Stacey AR, Dukes J, Baston E, Griffin S, Jakobsen BK, Vuidepot A, Harper S. TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex. Journal of Immunology. 2020;204:1943–1953. doi: 10.4049/jimmunol.1900915. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

