Feature selection revisited in the single-cell era
- PMID: 34847932
- PMCID: PMC8638336
- DOI: 10.1186/s13059-021-02544-3
Feature selection revisited in the single-cell era
Abstract
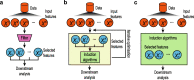
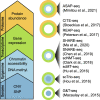
Recent advances in single-cell biotechnologies have resulted in high-dimensional datasets with increased complexity, making feature selection an essential technique for single-cell data analysis. Here, we revisit feature selection techniques and summarise recent developments. We review their application to a range of single-cell data types generated from traditional cytometry and imaging technologies and the latest array of single-cell omics technologies. We highlight some of the challenges and future directions and finally consider their scalability and make general recommendations on each type of feature selection method. We hope this review stimulates future research and application of feature selection in the single-cell era.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Guyon I, Elisseeff A. An introduction to variable and feature selection. Journal of Machine Learning Research. 2003;3:1157–1182.
-
- Lazar C, Taminau J, Meganck S, Steenhoff D, Coletta A, Molter C, de Schaetzen V, Duque R, Bersini H, Nowe A. A survey on filter techniques for feature selection in gene expression microarray analysis. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2012;9(4):1106–1119. doi: 10.1109/TCBB.2012.33. - DOI - PubMed
-
- Bolón-Canedo V, Sánchez-Marono N, Alonso-Betanzos A, Benítez JM, Herrera F. A review of microarray datasets and applied feature selection methods. Information Sciences. 2014;282:111–135. doi: 10.1016/j.ins.2014.05.042. - DOI