The Precision Interventions for Severe and/or Exacerbation-Prone (PrecISE) Asthma Network: An overview of Network organization, procedures, and interventions
- PMID: 34848210
- PMCID: PMC8821377
- DOI: 10.1016/j.jaci.2021.10.035
The Precision Interventions for Severe and/or Exacerbation-Prone (PrecISE) Asthma Network: An overview of Network organization, procedures, and interventions
Erratum in
-
Corrigendum.J Allergy Clin Immunol. 2022 Aug;150(2):491. doi: 10.1016/j.jaci.2022.06.001. J Allergy Clin Immunol. 2022. PMID: 35934682 Free PMC article. No abstract available.
Abstract
Asthma is a heterogeneous disease, with multiple underlying inflammatory pathways and structural airway abnormalities that impact disease persistence and severity. Recent progress has been made in developing targeted asthma therapeutics, especially for subjects with eosinophilic asthma. However, there is an unmet need for new approaches to treat patients with severe and exacerbation-prone asthma, who contribute disproportionately to disease burden. Extensive deep phenotyping has revealed the heterogeneous nature of severe asthma and identified distinct disease subtypes. A current challenge in the field is to translate new and emerging knowledge about different pathobiologic mechanisms in asthma into patient-specific therapies, with the ultimate goal of modifying the natural history of disease. Here, we describe the Precision Interventions for Severe and/or Exacerbation-Prone Asthma (PrecISE) Network, a groundbreaking collaborative effort of asthma researchers and biostatisticians from around the United States. The PrecISE Network was designed to conduct phase II/proof-of-concept clinical trials of precision interventions in the population with severe asthma, and is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health. Using an innovative adaptive platform trial design, the PrecISE Network will evaluate up to 6 interventions simultaneously in biomarker-defined subgroups of subjects. We review the development and organizational structure of the PrecISE Network, and choice of interventions being studied. We hope that the PrecISE Network will enhance our understanding of asthma subtypes and accelerate the development of therapeutics for severe asthma.
Keywords: Severe asthma; adaptive clinical trial design; asthma exacerbation; biomarker; non–type 2 asthma; patient advisory committee; precision medicine; type 2 asthma.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure of potential conflict of interest: S. N. Georas reports receiving grants from the National Institutes of Health (NIH) during the conduct of the study, and consulting fees from Merck outside of the submitted work. R. J. Wright reports grants from the NIH, during the conduct of the study. E. Israel reports grants from the NIH, Patient-Centered Outcomes Research Institute (PCORI), and Gossamer Bio; grants and nonfinancial support from Circassia; grants and personal fees from AstraZeneca, Avillion, and Novartis; personal fees and nonfinancial support from Genentech, GlaxoSmithKline (GSK), and Teva; personal fees from AB Science, Allergy and Asthma Network, Amgen, Arrowhead Pharmaceuticals, Biometry, Equillium, Merck, Pneuma Respiratory, the National Heart, Lung, and Blood Institute (NHLBI), PPS Health, Regeneron, Sanofi Genzyme, Sienna Biopharmaceutical, Teva, and Cowen; nonfinancial support from Boehringer Ingelheim; and other considerations from Vorso, outside the submitted work. L. M. LaVange reports grants from the NHLBI, during the conduct of the study. P. Akuthota reports grants from the NIH, during the conduct of the study; personal fees from WebMD/Medscape, AHK, Prime CME, UpToDate, Rockpointe, Projects in Knowledge, MJH LifeSciences, and Vindico CME, and grants and personal fees from GSK and AstraZeneca, outside the submitted work. T. F. Carr reports grants from the NHLBI, during the conduct of the study; grants and personal fees from AstraZeneca, Regeneron, and Novartis, and personal fees from GSK and Genentech, outside the submitted work. L. C. Denlinger reports grants from the NHLBI and American Lung Association / Asthma Clinical Research Centers, during the conduct of the study; and other considerations from AstraZeneca, Boehringer Ingelheim, Genentech, GSK, Sanofi-Genzyme-Regeneron, and Teva, outside the submitted work. M. L. Fajt reports personal fees from the American Academy of Allergy, Asthma & Immunology, and grants from Breathe PA Organization, outside the submitted work. W. Phipatanakul reports grants from Boehringer Ingelheim, and grants and personal fees from Genentech, Sanofi Regeneron, AstraZeneca, GlaxoSmithKline, Genzyme, and Teva, outside the submitted work. S. J. Szefler reports grants from Propeller Health, and other considerations from Moderna, GSK, AstraZeneca, Sanofi, and Regeneron, outside the submitted work. L. B. Bacharier reports grants from the NIH/NHLBI, during the conduct of the study; personal fees from GSK, Genentech/Novartis, DBV Technologies, AstraZeneca, WebMD/Medscape, Sanofi/Regeneron, Vectura, and Circassia, and personal fees and nonfinancial support from Merck, Teva, and Boehringer Ingelheim, outside the submitted work. M. Castro reports grants from the NIH, ALA, PCORI, Pulmatrix, and Shionogi; grants and personal fees from AstraZeneca, Novartis, GSK, and Sanofi-Aventis; and personal fees from Teva, Genentech, and Regeneron, outside the submitted work. J. C. Cardet reports personal fees from AstraZeneca, GSK, and Genentech, outside the submitted work. S. A. A. Comhair reports grants from the NHLBI, during the conduct of the study, and a patent, “Capric Acid and Myristic Acid Compositions for Treatment Conditions (publication no. 20200230094),” pending. R. A. Covar reports grants from the NHLBI, during the conduct of the study; grants from Avillion Bond, GSK, and American Lung Association/Asthma Clinical Research Centers (ALA/ACRC), and personal fees from Sanofi Regeneron, outside the submitted work. E. A. DiMango reports personal fees from AstraZeneca, outside the submitted work. K. Erwin reports grants from the NIH and the Robert Wood Johnson Foundation, grants and personal fees from the Arnold Foundation, and personal fees from the National Academy of Medicine and IIT Institute of Design, outside the submitted work. S. C. Erzurum reports grants from the NIH, during the conduct of the study. J. V. Fahy reports personal fees from Boehringer Ingelheim, Pieris, Arrowhead Pharmaceuticals, Gossamer, Ionis Pharmaceuticals, and Suzhou Connect Biopharmaceuticals, Ltd, outside the submitted work, and has a patent WO2014153009A2, “Thiosaccharide mucolytic agents,” issued, and a patent WO2017197360, “CT Mucus Score—A new scoring system that quantifies airway mucus impaction using CT lung scans,” pending. B. Gaston reports personal fees from Laurel, and other considerations from Respiratory Research, Inc, outside the submitted work. L. B. Gerald reports grants from the NIH, the Banner Health Foundation, the American Lung Association, and Thayer Medical Corporation; and personal fees from UpToDate, Wolters Kluwer, the University of Illinois, and the Nemours Foundation, outside the submitted work. E. A. Hoffman is a founder and shareholder of VIDA Diagnostics, a company commercializing lung image analysis software developed, in part, at the University of Iowa. D. J. Jackson reports grants from the NIH-Environmental Influences on Child Health Outcomes (ECHO)/Children’s Respiratory Research Workgroup (CREW), the NHLBI, and the National Institute of Allergy and Infectious Diseases (NIAID), during the conduct of the study; personal fees from Sanofi Regeneron, Pfizer, AstraZeneca, Novartis, and Vifor Pharma, and grants and personal fees from GlaxoSmithKline, outside the submitted work. N. N. Jarjour reports grants from the NIH-NHLBI during the conduct of the study; grants and personal fees from AstraZeneca, and personal fees from GSK and Boehringer Ingelheim for consultations outside the submitted work. M. Kraft reports grants from the NIH and the ALA; grants and personal fees from Sanofi Regeneron, Chiesi Pharmaceuticals, and AstraZeneca; and personal fees from Genentech, outside the submitted work. J. A. Krishnan reports grants from the NIH-NHLBI during the conduct of the study, grants from Regeneron, the COPD Foundation, and Sergey Brin Family Foundation, and personal fees from GSK, outside the submitted work. M. C. Liu reports grants from the NIH, during the conduct of the study; personal fees from UpToDate, grants and personal fees from Gossamer Bio and GSK, and other considerations from AstraZeneca, outside the submitted work. F. Martinez reports grants from the NIH/NHLBI, NIH/National Institute of Environmental Health Sciences, NIH/NIAID, and NIH/Office of Director, outside the submitted work. W. C. Moore reports grants from the NHLBI during the conduct of the study; grants and personal fees from AstraZeneca, GSK, and Sanofi Regeneron, and grants from Gossamer Bio, Inc, Cumberland Pharmaceuticals, and Genentech, outside the submitted work. V. E. Ortega reports personal fees from Regeneron and Sanofi, outside the submitted work. M. C. Peters reports grants from AstraZeneca, Boehringer Ingelheim, Genentech, GSK, Sanofi-Genzyme-Regeneron, and Teva, and personal fees from OrbiMed, outside the submitted work. K. Ross reports grants from the NIH, during the conduct of the study; grants and nonfinancial support from Teva; and grants from AstraZeneca and Novartis. L. J. Smith reports grants from the NHLBI, during the conduct of the study. M. E. Wechsler reports grants from the Jin Hua Foundation; grants, personal fees, and nonfinancial support from Teva; grants and personal fees from Sanofi, Regeneron, and Cohero Health; and personal fees from Amgen, GSK, Genentech, Novartis, Sentien, Restorbio, Equillium, Genzyme, Pulmatrix, Cytoreason, Cerecor, Incyte, Sound Biologics, Kinaset, Boehringer Ingelheim, and AstraZeneca, outside the submitted work. S. E. Wenzel reports grants from Teva; personal fees from Sanofi; and grants and personal fees from AstraZeneca, Knopp, and Novartis, outside the submitted work. S. R. White reports grants from the NHLBI, during the conduct of the study; and personal fees from Sanofi Regeneron and AstraZeneca, outside the submitted work. A. A. Zeki reports grants from InVixa, Inc; fees from Sanofi Regeneron and the Center for Healthcare Innovation (cIMBA); other considerations from InStatin, Inc, and InVixa, Inc, outside the submitted work; and has a patent PCT/US2020/025543 pending (no sponsor) and a Provisional Patent Application: Serial No. 63/158-144. UC 2020-551-2; 052564-563P02US. The rest of the authors declare that they have no relevant conflicts of interest.
Figures



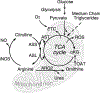

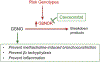

References
-
- Israel E, Reddel HK. Severe and difficult-to-treat asthma in adults. N Engl J Med 2017;377:965–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UG1 HL139124/HL/NHLBI NIH HHS/United States
- K23 AI135094/AI/NIAID NIH HHS/United States
- R03 AI139648/AI/NIAID NIH HHS/United States
- P30 ES001247/ES/NIEHS NIH HHS/United States
- K23 AI125785/AI/NIAID NIH HHS/United States
- UL1 TR001422/TR/NCATS NIH HHS/United States
- P30 ES023513/ES/NIEHS NIH HHS/United States
- UG1 HL139119/HL/NHLBI NIH HHS/United States
- R03 HL148486/HL/NHLBI NIH HHS/United States
- UL1 TR002389/TR/NCATS NIH HHS/United States
- UL1 TR002373/TR/NCATS NIH HHS/United States
- UG1 HL139118/HL/NHLBI NIH HHS/United States
- UG1 HL139117/HL/NHLBI NIH HHS/United States
- UG1 HL139125/HL/NHLBI NIH HHS/United States
- UL1 TR002366/TR/NCATS NIH HHS/United States
- UG1 HL139106/HL/NHLBI NIH HHS/United States
- UL1 TR000427/TR/NCATS NIH HHS/United States
- U24 HL138998/HL/NHLBI NIH HHS/United States
- UL1 TR001857/TR/NCATS NIH HHS/United States
- UL1 TR001872/TR/NCATS NIH HHS/United States
- P30 ES005605/ES/NIEHS NIH HHS/United States
- U24 TR001608/TR/NCATS NIH HHS/United States
- UG1 HL139123/HL/NHLBI NIH HHS/United States
- UL1 TR001442/TR/NCATS NIH HHS/United States
- T32 AI007512/AI/NIAID NIH HHS/United States
- UL1 TR002489/TR/NCATS NIH HHS/United States
- U54 GM104940/GM/NIGMS NIH HHS/United States
- P50 CA058223/CA/NCI NIH HHS/United States
- UG1 HL139098/HL/NHLBI NIH HHS/United States
- R01 HL148715/HL/NHLBI NIH HHS/United States
- U01 AI097073/AI/NIAID NIH HHS/United States
- UG1 HL139126/HL/NHLBI NIH HHS/United States
- L30 HL149106/HL/NHLBI NIH HHS/United States
- UG1 HL139054/HL/NHLBI NIH HHS/United States

