Ribosome profiling reveals novel regulation of C9ORF72 GGGGCC repeat-containing RNA translation
- PMID: 34848561
- PMCID: PMC8906550
- DOI: 10.1261/rna.078963.121
Ribosome profiling reveals novel regulation of C9ORF72 GGGGCC repeat-containing RNA translation
Abstract
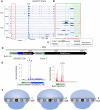
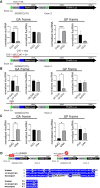
GGGGCC (G4C2) repeat expansion in the first intron of C9ORF72 causes amyotrophic lateral sclerosis and frontotemporal dementia. Repeat-containing RNA is translated into dipeptide repeat (DPR) proteins, some of which are neurotoxic. Using dynamic ribosome profiling, we identified three translation initiation sites in the intron upstream of (G4C2) repeats; these sites are detected irrespective of the presence or absence of the repeats. During translocation, ribosomes appear to be stalled on the repeats. An AUG in the preceding C9ORF72 exon initiates a uORF that inhibits downstream translation. Polysome isolation indicates that unspliced (G4C2) repeat-containing RNA is a substrate for DPR protein synthesis. (G4C2) repeat-containing RNA translation is 5' cap-independent but inhibited by the initiation factor DAP5, suggesting an interplay with uORF function. These results define novel translational mechanisms of expanded (G4C2) repeat-containing RNA in disease.
Keywords: ALS; C9ORF72; DAP5; frontotemporal dementia; ribosome profiling; ribosome stalling; uORF translation.
© 2022 van ‘t Spijker et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Almeida S, Zhang Z, Coppola G, Mao W, Futai K, Karydas A, Geschwind MD, Tartaglia MC, Gao F, Gianni D, et al. 2012. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep 2: 789–798. 10.1016/j.celrep.2012.09.007 - DOI - PMC - PubMed
-
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. 2013. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol 126: 385. 10.1007/s00401-013-1149-y - DOI - PMC - PubMed
-
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646. 10.1016/j.neuron.2013.02.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
