Regulatory circuits involving bud dormancy factor PpeDAM6
- PMID: 34848702
- PMCID: PMC8632999
- DOI: 10.1038/s41438-021-00706-9
Regulatory circuits involving bud dormancy factor PpeDAM6
Abstract
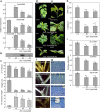
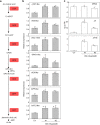
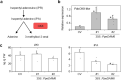
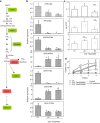
DORMANCY-ASSOCIATED MADS-BOX (DAM) genes have recently emerged as key potential regulators of the dormancy cycle and climate adaptation in perennial species. Particularly, PpeDAM6 has been proposed to act as a major repressor of bud dormancy release and bud break in peach (Prunus persica). PpeDAM6 expression is downregulated concomitantly with the perception of a given genotype-dependent accumulation of winter chilling time, and the coincident enrichment in H3K27me3 chromatin modification at a specific genomic region. We have identified three peach BASIC PENTACYSTEINE PROTEINs (PpeBPCs) interacting with two GA-repeat motifs present in this H3K27me3-enriched region. Moreover, PpeBPC1 represses PpeDAM6 promoter activity by transient expression experiments. On the other hand, the heterologous overexpression of PpeDAM6 in European plum (Prunus domestica) alters plant vegetative growth, resulting in dwarf plants tending toward shoot meristem collapse. These alterations in vegetative growth of transgenic lines associate with impaired hormone homeostasis due to the modulation of genes involved in jasmonic acid, cytokinin, abscisic acid, and gibberellin pathways, and the downregulation of shoot meristem factors, specifically in transgenic leaf and apical tissues. The expression of many of these genes is also modified in flower buds of peach concomitantly with PpeDAM6 downregulation, which suggests a role of hormone homeostasis mechanisms in PpeDAM6-dependent maintenance of floral bud dormancy and growth repression.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Singh RK, Svystun T, AlDahmash B, Jönsson AM, Bhalerao RP. Photoperiod- and temperature-mediated control of phenology in trees – a molecular perspective. N. Phytol. 2017;213:511–524. - PubMed
-
- Heide OM, Prestrud AK. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol. 2005;25:109–114. - PubMed
-
- Cooke JEK, Eriksson ME, Junttila O. The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant Cell Environ. 2012;35:1707–1728. - PubMed

