Genome-wide analysis of acute low salinity tolerance in the eastern oyster Crassostrea virginica and potential of genomic selection for trait improvement
- PMID: 34849774
- PMCID: PMC8727987
- DOI: 10.1093/g3journal/jkab368
Genome-wide analysis of acute low salinity tolerance in the eastern oyster Crassostrea virginica and potential of genomic selection for trait improvement
Abstract
As the global demand for seafood increases, research into the genetic basis of traits that can increase aquaculture production is critical. The eastern oyster (Crassostrea virginica) is an important aquaculture species along the Atlantic and Gulf Coasts of the United States, but increases in heavy rainfall events expose oysters to acute low salinity conditions, which negatively impact production. Low salinity survival is known to be a moderately heritable trait, but the genetic architecture underlying this trait is still poorly understood. In this study, we used ddRAD sequencing to generate genome-wide single-nucleotide polymorphism (SNP) data for four F2 families to investigate the genomic regions associated with survival in extreme low salinity (<3). SNP data were also used to assess the feasibility of genomic selection (GS) for improving this trait. Quantitative trait locus (QTL) mapping and combined linkage disequilibrium analysis revealed significant QTL on eastern oyster chromosomes 1 and 7 underlying both survival and day to death in a 36-day experimental challenge. Significant QTL were located in genes related to DNA/RNA function and repair, ion binding and membrane transport, and general response to stress. GS was investigated using Bayesian linear regression models and prediction accuracies ranged from 0.48 to 0.57. Genomic prediction accuracies were largest using the BayesB prior and prediction accuracies did not substantially decrease when SNPs located within the QTL region on Chr1 were removed, suggesting that this trait is controlled by many genes of small effect. Our results suggest that GS will likely be a viable option for improvement of survival in extreme low salinity.
Keywords: Crassostrea virginica; BGLR; QTL mapping; genomic selection; oyster aquaculture; salinity tolerance.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures
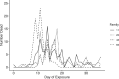


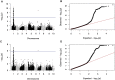
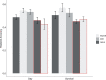
References
-
- Allen SK, Small JM, Kube PD.. 2021. Genetic parameters for Crassostrea virginica and their application to family-based breeding in the mid-Atlantic, USA. Aquaculture. 538:736578.
-
- Andrews JD, Quayle DB, Haven D.. 1959. Fresh-water kill of oysters (Crassostrea virginica) in James River, Virginia, 1958. Proc Natl Shellfish Assoc. 49:29–49.
-
- Barría A, Trinh TQ, Mahmuddin M, Benzie JAH, Chadag VM, et al.2020. Genetic parameters for resistance to Tilapia Lake Virus (TiLV) in Nile tilapia (Oreochromis niloticus). Aquaculture. 522:735126.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
