Enhancing biological signals and detection rates in single-cell RNA-seq experiments with cDNA library equalization
- PMID: 34850101
- PMCID: PMC8789062
- DOI: 10.1093/nar/gkab1071
Enhancing biological signals and detection rates in single-cell RNA-seq experiments with cDNA library equalization
Abstract
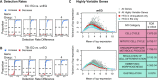
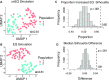
Considerable effort has been devoted to refining experimental protocols to reduce levels of technical variability and artifacts in single-cell RNA-sequencing data (scRNA-seq). We here present evidence that equalizing the concentration of cDNA libraries prior to pooling, a step not consistently performed in single-cell experiments, improves gene detection rates, enhances biological signals, and reduces technical artifacts in scRNA-seq data. To evaluate the effect of equalization on various protocols, we developed Scaffold, a simulation framework that models each step of an scRNA-seq experiment. Numerical experiments demonstrate that equalization reduces variation in sequencing depth and gene-specific expression variability. We then performed a set of experiments in vitro with and without the equalization step and found that equalization increases the number of genes that are detected in every cell by 17-31%, improves discovery of biologically relevant genes, and reduces nuisance signals associated with cell cycle. Further support is provided in an analysis of publicly available data.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Svensson V., Vento-Tormo R., Teichmann S.A.. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018; 13:599–604. - PubMed
-
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M.et al.. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015; 16:278. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

