PharmacoDB 2.0: improving scalability and transparency of in vitro pharmacogenomics analysis
- PMID: 34850112
- PMCID: PMC8728279
- DOI: 10.1093/nar/gkab1084
PharmacoDB 2.0: improving scalability and transparency of in vitro pharmacogenomics analysis
Abstract
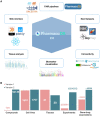
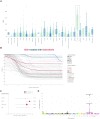
Cancer pharmacogenomics studies provide valuable insights into disease progression and associations between genomic features and drug response. PharmacoDB integrates multiple cancer pharmacogenomics datasets profiling approved and investigational drugs across cell lines from diverse tissue types. The web-application enables users to efficiently navigate across datasets, view and compare drug dose-response data for a specific drug-cell line pair. In the new version of PharmacoDB (version 2.0, https://pharmacodb.ca/), we present (i) new datasets such as NCI-60, the Profiling Relative Inhibition Simultaneously in Mixtures (PRISM) dataset, as well as updated data from the Genomics of Drug Sensitivity in Cancer (GDSC) and the Genentech Cell Line Screening Initiative (gCSI); (ii) implementation of FAIR data pipelines using ORCESTRA and PharmacoDI; (iii) enhancements to drug-response analysis such as tissue distribution of dose-response metrics and biomarker analysis; and (iv) improved connectivity to drug and cell line databases in the community. The web interface has been rewritten using a modern technology stack to ensure scalability and standardization to accommodate growing pharmacogenomics datasets. PharmacoDB 2.0 is a valuable tool for mining pharmacogenomics datasets, comparing and assessing drug-response phenotypes of cancer models.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Shoemaker R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006; 6:813–823. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

