MLL1 is regulated by KSHV LANA and is important for virus latency
- PMID: 34850113
- PMCID: PMC8682764
- DOI: 10.1093/nar/gkab1094
MLL1 is regulated by KSHV LANA and is important for virus latency
Abstract
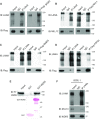
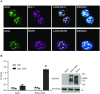
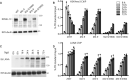
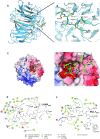
Mixed lineage leukemia 1 (MLL1) is a histone methyltransferase. Kaposi's sarcoma-associated herpesvirus (KSHV) is a leading cause of malignancy in AIDS. KSHV latently infects tumor cells and its genome is decorated with epigenetic marks. Here, we show that KSHV latency-associated nuclear antigen (LANA) recruits MLL1 to viral DNA where it establishes H3K4me3 modifications at the extensive KSHV terminal repeat elements during primary infection. LANA interacts with MLL1 complex members, including WDR5, integrates into the MLL1 complex, and regulates MLL1 activity. We describe the 1.5-Å crystal structure of N-terminal LANA peptide complexed with MLL1 complex member WDR5, which reveals a potential regulatory mechanism. Disruption of MLL1 expression rendered KSHV latency establishment highly deficient. This deficiency was rescued by MLL1 but not by catalytically inactive MLL1. Therefore, MLL1 is LANA regulable and exerts a central role in virus infection. These results suggest broad potential for MLL1 regulation, including by non-host factors.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Site-specific association with host and viral chromatin by Kaposi's sarcoma-associated herpesvirus LANA and its reversal during lytic reactivation.J Virol. 2014 Jun;88(12):6762-77. doi: 10.1128/JVI.00268-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696474 Free PMC article.
-
Full-Length Isoforms of Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Accumulate in the Cytoplasm of Cells Undergoing the Lytic Cycle of Replication.J Virol. 2017 Nov 30;91(24):e01532-17. doi: 10.1128/JVI.01532-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978712 Free PMC article.
-
Activated Nrf2 Interacts with Kaposi's Sarcoma-Associated Herpesvirus Latency Protein LANA-1 and Host Protein KAP1 To Mediate Global Lytic Gene Repression.J Virol. 2015 Aug;89(15):7874-92. doi: 10.1128/JVI.00895-15. Epub 2015 May 20. J Virol. 2015. PMID: 25995248 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
Cited by
-
Molecular mechanisms of viral oncogenesis in haematological malignancies: perspectives from metabolic reprogramming, epigenetic regulation and immune microenvironment remodeling.Exp Hematol Oncol. 2025 May 10;14(1):69. doi: 10.1186/s40164-025-00655-2. Exp Hematol Oncol. 2025. PMID: 40349096 Free PMC article. Review.
-
Kaposi's sarcoma herpesvirus latency-associated nuclear antigen broadly regulates viral gene expression and is essential for lytic infection.PLoS Pathog. 2024 Jan 17;20(1):e1011907. doi: 10.1371/journal.ppat.1011907. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38232124 Free PMC article.
-
The NTE domain of PTENα/β promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.Cell Death Dis. 2024 May 14;15(5):335. doi: 10.1038/s41419-024-06714-6. Cell Death Dis. 2024. PMID: 38744853 Free PMC article.
-
LANA-Dependent Transcription-Replication Conflicts and R-Loops at the Terminal Repeats (TR) Correlate with KSHV Episome Maintenance.bioRxiv [Preprint]. 2025 Mar 13:2025.03.10.642343. doi: 10.1101/2025.03.10.642343. bioRxiv. 2025. PMID: 40161765 Free PMC article. Preprint.
-
Kaposi's sarcoma-associated herpesvirus (KSHV) LANA prevents KSHV episomes from degradation.J Virol. 2024 Feb 20;98(2):e0126823. doi: 10.1128/jvi.01268-23. Epub 2024 Jan 19. J Virol. 2024. PMID: 38240588 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

