Expression Atlas update: gene and protein expression in multiple species
- PMID: 34850121
- PMCID: PMC8728300
- DOI: 10.1093/nar/gkab1030
Expression Atlas update: gene and protein expression in multiple species
Abstract
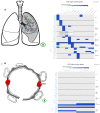
The EMBL-EBI Expression Atlas is an added value knowledge base that enables researchers to answer the question of where (tissue, organism part, developmental stage, cell type) and under which conditions (disease, treatment, gender, etc) a gene or protein of interest is expressed. Expression Atlas brings together data from >4500 expression studies from >65 different species, across different conditions and tissues. It makes these data freely available in an easy to visualise form, after expert curation to accurately represent the intended experimental design, re-analysed via standardised pipelines that rely on open-source community developed tools. Each study's metadata are annotated using ontologies. The data are re-analyzed with the aim of reproducing the original conclusions of the underlying experiments. Expression Atlas is currently divided into Bulk Expression Atlas and Single Cell Expression Atlas. Expression Atlas contains data from differential studies (microarray and bulk RNA-Seq) and baseline studies (bulk RNA-Seq and proteomics), whereas Single Cell Expression Atlas is currently dedicated to Single Cell RNA-Sequencing (scRNA-Seq) studies. The resource has been in continuous development since 2009 and it is available at https://www.ebi.ac.uk/gxa.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 108437/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/P024599/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 221401/Z/20/Z /WT_/Wellcome Trust/United Kingdom
- BB/T019670/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

