Enzymatic deamination of the epigenetic nucleoside N6-methyladenosine regulates gene expression
- PMID: 34850126
- PMCID: PMC8643624
- DOI: 10.1093/nar/gkab1124
Enzymatic deamination of the epigenetic nucleoside N6-methyladenosine regulates gene expression
Abstract
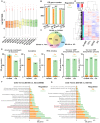
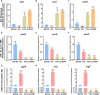
N6-methyladenosine (m6A) modification is the most extensively studied epigenetic modification due to its crucial role in regulating an array of biological processes. Herein, Bsu06560, formerly annotated as an adenine deaminase derived from Bacillus subtilis 168, was recognized as the first enzyme capable of metabolizing the epigenetic nucleoside N6-methyladenosine. A model of Bsu06560 was constructed, and several critical residues were putatively identified via mutational screening. Two mutants, F91L and Q150W, provided a superiorly enhanced conversion ratio of adenosine and N6-methyladenosine. The CRISPR-Cas9 system generated Bsu06560-knockout, F91L, and Q150W mutations from the B. subtilis 168 genome. Transcriptional profiling revealed a higher global gene expression level in BS-F91L and BS-Q150W strains with enhanced N6-methyladenosine deaminase activity. The differentially expressed genes were categorized using GO, COG, KEGG and verified through RT-qPCR. This study assessed the crucial roles of Bsu06560 in regulating adenosine and N6-methyladenosine metabolism, which influence a myriad of biological processes. This is the first systematic research to identify and functionally annotate an enzyme capable of metabolizing N6-methyladenosine and highlight its significant roles in regulation of bacterial metabolism. Besides, this study provides a novel method for controlling gene expression through the mutations of critical residues.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
The C-terminal structure of the N6-methyladenosine deaminase YerA and its role in deamination.Biochem J. 2025 Feb 12;482(4):BCJ20240728. doi: 10.1042/BCJ20240728. Biochem J. 2025. PMID: 39876819 Free PMC article.
-
Epigenetic N6-methyladenosine modification of RNA and DNA regulates cancer.Cancer Biol Med. 2020 Feb 15;17(1):9-19. doi: 10.20892/j.issn.2095-3941.2019.0347. Cancer Biol Med. 2020. PMID: 32296573 Free PMC article. Review.
-
Epigenetic regulation of mRNA N6-methyladenosine modifications in mammalian gametogenesis.Mol Hum Reprod. 2021 May 8;27(5):gaab025. doi: 10.1093/molehr/gaab025. Mol Hum Reprod. 2021. PMID: 33823008 Review.
-
Epigenetic Methylations on N6-Adenine and N6-Adenosine with the same Input but Different Output.Int J Mol Sci. 2019 Jun 15;20(12):2931. doi: 10.3390/ijms20122931. Int J Mol Sci. 2019. PMID: 31208067 Free PMC article. Review.
-
[Progress in epigenetic modification of mRNA and the function of m6A modification].Sheng Wu Gong Cheng Xue Bao. 2019 May 25;35(5):775-783. doi: 10.13345/j.cjb.180416. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 31222996 Review. Chinese.
Cited by
-
Biocontrol potential of Bacillus subtilis CTXW 7-6-2 against kiwifruit soft rot pathogens revealed by whole-genome sequencing and biochemical characterisation.Front Microbiol. 2022 Dec 1;13:1069109. doi: 10.3389/fmicb.2022.1069109. eCollection 2022. Front Microbiol. 2022. PMID: 36532498 Free PMC article.
-
A model industrial workhorse: Bacillus subtilis strain 168 and its genome after a quarter of a century.Microb Biotechnol. 2023 Jun;16(6):1203-1231. doi: 10.1111/1751-7915.14257. Epub 2023 Apr 1. Microb Biotechnol. 2023. PMID: 37002859 Free PMC article. Review.
-
Structure Characterization of Escherichia coli Pseudouridine Kinase PsuK.Front Microbiol. 2022 Jun 17;13:926099. doi: 10.3389/fmicb.2022.926099. eCollection 2022. Front Microbiol. 2022. PMID: 35783380 Free PMC article.
-
The C-terminal structure of the N6-methyladenosine deaminase YerA and its role in deamination.Biochem J. 2025 Feb 12;482(4):BCJ20240728. doi: 10.1042/BCJ20240728. Biochem J. 2025. PMID: 39876819 Free PMC article.
-
OpDetect: A convolutional and recurrent neural network classifier for precise and sensitive operon detection from RNA-seq data.PLoS One. 2025 Aug 1;20(8):e0329355. doi: 10.1371/journal.pone.0329355. eCollection 2025. PLoS One. 2025. PMID: 40748965 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

