Discovery of highly reactive self-splicing group II introns within the mitochondrial genomes of human pathogenic fungi
- PMID: 34850132
- PMCID: PMC8643640
- DOI: 10.1093/nar/gkab1077
Discovery of highly reactive self-splicing group II introns within the mitochondrial genomes of human pathogenic fungi
Abstract
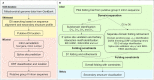
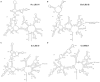
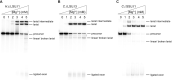
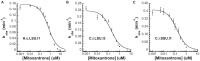
Fungal pathogens represent an expanding global health threat for which treatment options are limited. Self-splicing group II introns have emerged as promising drug targets, but their development has been limited by a lack of information on their distribution and architecture in pathogenic fungi. To meet this challenge, we developed a bioinformatic workflow for scanning sequence data to identify unique RNA structural signatures within group II introns. Using this approach, we discovered a set of ubiquitous introns within thermally dimorphic fungi (genera of Blastomyces, Coccidioides and Histoplasma). These introns are the most biochemically reactive group II introns ever reported, and they self-splice rapidly under near-physiological conditions without protein cofactors. Moreover, we demonstrated the small molecule targetability of these introns by showing that they can be inhibited by the FDA-approved drug mitoxantrone in vitro. Taken together, our results highlight the utility of structure-based informatic searches for identifying riboregulatory elements in pathogens, revealing a striking diversity of reactive self-splicing introns with great promise as antifungal drug targets.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Highly Reactive Group I Introns Ubiquitous in Pathogenic Fungi.J Mol Biol. 2024 Apr 15;436(8):168513. doi: 10.1016/j.jmb.2024.168513. Epub 2024 Mar 5. J Mol Biol. 2024. PMID: 38447889
-
Molecular evolution of the fungi: human pathogens.Mol Biol Evol. 1992 Sep;9(5):893-904. doi: 10.1093/oxfordjournals.molbev.a040766. Mol Biol Evol. 1992. PMID: 1528111
-
The Agaricus bisporus cox1 gene: the longest mitochondrial gene and the largest reservoir of mitochondrial group i introns.PLoS One. 2010 Nov 18;5(11):e14048. doi: 10.1371/journal.pone.0014048. PLoS One. 2010. PMID: 21124976 Free PMC article.
-
Thermally Dimorphic Human Fungal Pathogens--Polyphyletic Pathogens with a Convergent Pathogenicity Trait.Cold Spring Harb Perspect Med. 2014 Nov 10;5(8):a019794. doi: 10.1101/cshperspect.a019794. Cold Spring Harb Perspect Med. 2014. PMID: 25384771 Free PMC article. Review.
-
Group II introns: elaborate ribozymes.Biochimie. 1996;78(6):474-87. doi: 10.1016/0300-9084(96)84754-7. Biochimie. 1996. PMID: 8915537 Review.
Cited by
-
A molecular beacon assay for monitoring RNA splicing.Nucleic Acids Res. 2022 Jul 22;50(13):e74. doi: 10.1093/nar/gkac242. Nucleic Acids Res. 2022. PMID: 35438748 Free PMC article.
-
Protein-free catalysis of DNA hydrolysis and self-integration by a ribozyme.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1224. doi: 10.1093/nar/gkae1224. Nucleic Acids Res. 2025. PMID: 39698822 Free PMC article.
-
Uncovering the role of mitochondrial genome in pathogenicity and drug resistance in pathogenic fungi.Front Cell Infect Microbiol. 2025 Apr 16;15:1576485. doi: 10.3389/fcimb.2025.1576485. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40308969 Free PMC article. Review.
-
Development and comprehensive evaluation of scarless circularization systems for circular RNA therapeutics.Mol Ther Nucleic Acids. 2025 Jun 9;36(3):102587. doi: 10.1016/j.omtn.2025.102587. eCollection 2025 Sep 9. Mol Ther Nucleic Acids. 2025. PMID: 40606646 Free PMC article.
-
Computational De Novo Design of Group II Introns Yields Highly Active Ribozymes.Chembiochem. 2025 Jul 18;26(14):e202500356. doi: 10.1002/cbic.202500356. Epub 2025 Jun 30. Chembiochem. 2025. PMID: 40504414 Free PMC article.
References
-
- Fisher M.C., Hawkins N.J., Sanglard D., Gurr S.J.. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018; 360:739–742. - PubMed
-
- Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., White T.C.. Hidden killers: human fungal infections. Sci. Transl. Med. 2012; 4:165rv113. - PubMed
-
- Knox K.S. Perspective on coccidioidomycosis and histoplasmosis. Am. J. Respir. Crit. Care Med. 2014; 189:752–753. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

