PRODORIC: state-of-the-art database of prokaryotic gene regulation
- PMID: 34850133
- PMCID: PMC8728284
- DOI: 10.1093/nar/gkab1110
PRODORIC: state-of-the-art database of prokaryotic gene regulation
Abstract
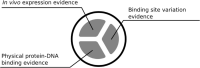
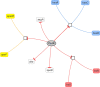
PRODORIC is worldwide one of the largest collections of prokaryotic transcription factor binding sites from multiple bacterial sources with corresponding interpretation and visualization tools. With the introduction of PRODORIC2 in 2017, the transition to a modern web interface and maintainable backend was started. With this latest PRODORIC release the database backend is now fully API-based and provides programmatical access to the complete PRODORIC data. The visualization tools Genome Browser and ProdoNet from the original PRODORIC have been reintroduced and were integrated into the PRODORIC website. Missing input and output options from the original Virtual Footprint were added again for position weight matrix pattern-based searches. The whole PRODORIC dataset was reannotated. Every transcription factor binding site was re-evaluated to increase the overall database quality. During this process, additional parameters, like bound effectors, regulation type and different types of experimental evidence have been added for every transcription factor. Additionally, 109 new transcription factors and 6 new organisms have been added. PRODORIC is publicly available at https://www.prodoric.de.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

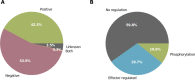


References
-
- Abril A.G., Rama J.L.R., Sánchez-Pérez A., Villa T.G.. Prokaryotic sigma factors and their transcriptional counterparts in Archaea and Eukarya. Appl. Microbiol. Biot. 2020; 104:4289–4302. - PubMed
-
- Griesenbeck J., Tschochner H., Grohmann D.. Structure and function of RNA polymerases and the transcription machineries. Macromol. Protein Complexes. 2017; 225–270. - PubMed
-
- Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. - PubMed
-
- Davis M.C., Kesthely C.A., Franklin E.A., MacLellan S.R.. The essential activities of the bacterial sigma factor. Can. J. Microbiol. 2017; 63:89–99. - PubMed
-
- Laudet V. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J. Mol. Endocrinol. 1997; 19:207–226. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

