DisProt in 2022: improved quality and accessibility of protein intrinsic disorder annotation
- PMID: 34850135
- PMCID: PMC8728214
- DOI: 10.1093/nar/gkab1082
DisProt in 2022: improved quality and accessibility of protein intrinsic disorder annotation
Abstract
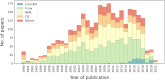
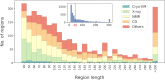
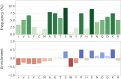
The Database of Intrinsically Disordered Proteins (DisProt, URL: https://disprot.org) is the major repository of manually curated annotations of intrinsically disordered proteins and regions from the literature. We report here recent updates of DisProt version 9, including a restyled web interface, refactored Intrinsically Disordered Proteins Ontology (IDPO), improvements in the curation process and significant content growth of around 30%. Higher quality and consistency of annotations is provided by a newly implemented reviewing process and training of curators. The increased curation capacity is fostered by the integration of DisProt with APICURON, a dedicated resource for the proper attribution and recognition of biocuration efforts. Better interoperability is provided through the adoption of the Minimum Information About Disorder (MIADE) standard, an active collaboration with the Gene Ontology (GO) and Evidence and Conclusion Ontology (ECO) consortia and the support of the ELIXIR infrastructure.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Romero P., Obradovic Z., Kissinger C.R., Villafranca J.E., Garner E., Guilliot S., Dunker A.K.. Thousands of proteins likely to have long disordered regions. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 1998; 1998:437–448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

