CRISPRroots: on- and off-target assessment of RNA-seq data in CRISPR-Cas9 edited cells
- PMID: 34850137
- PMCID: PMC8887420
- DOI: 10.1093/nar/gkab1131
CRISPRroots: on- and off-target assessment of RNA-seq data in CRISPR-Cas9 edited cells
Abstract
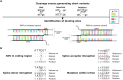
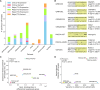
The CRISPR-Cas9 genome editing tool is used to study genomic variants and gene knockouts, and can be combined with transcriptomic analyses to measure the effects of such alterations on gene expression. But how can one be sure that differential gene expression is due to a successful intended edit and not to an off-target event, without performing an often resource-demanding genome-wide sequencing of the edited cell or strain? To address this question we developed CRISPRroots: CRISPR-Cas9-mediated edits with accompanying RNA-seq data assessed for on-target and off-target sites. Our method combines Cas9 and guide RNA binding properties, gene expression changes, and sequence variants between edited and non-edited cells to discover potential off-targets. Applied on seven public datasets, CRISPRroots identified critical off-target candidates that were overlooked in all of the corresponding previous studies. CRISPRroots is available via https://rth.dk/resources/crispr.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Doudna J.A., Charpentier E.. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014; 346:1258096. - PubMed
-
- Chuai G., Wang Q.-L., Liu Q.. In silico meets in vivo : towards computational CRISPR-based sgRNA design. Trends Biotechnol. 2017; 35:12–21. - PubMed
-
- Zischewski J., Fischer R., Bortesi L.. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017; 35:95–104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

