7',5'-alpha-bicyclo-DNA: new chemistry for oligonucleotide exon splicing modulation therapy
- PMID: 34850138
- PMCID: PMC8643641
- DOI: 10.1093/nar/gkab1097
7',5'-alpha-bicyclo-DNA: new chemistry for oligonucleotide exon splicing modulation therapy
Abstract
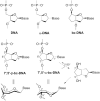
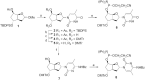
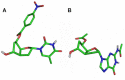
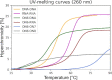
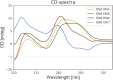
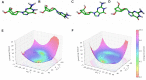
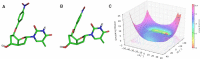
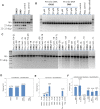
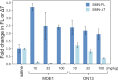
Antisense oligonucleotides are small pieces of modified DNA or RNA, which offer therapeutic potential for many diseases. We report on the synthesis of 7',5'-α-bc-DNA phosphoramidite building blocks, bearing the A, G, T and MeC nucleobases. Solid-phase synthesis was performed to construct five oligodeoxyribonucleotides containing modified thymidine residues, as well as five fully modified oligonucleotides. Incorporations of the modification inside natural duplexes resulted in strong destabilizing effects. However, fully modified strands formed very stable duplexes with parallel RNA complements. In its own series, 7',5'-α-bc-DNA formed duplexes with a surprising high thermal stability. CD spectroscopy and extensive molecular modeling indicated the adoption by the homo-duplex of a ladder-like structure, while hetero-duplexes with DNA or RNA still form helical structure. The biological properties of this new modification were investigated in animal models for Duchenne muscular dystrophy and spinal muscular atrophy, where exon splicing modulation can restore production of functional proteins. It was found that the 7',5'-α-bc-DNA scaffold confers a high biostability and a good exon splicing modulation activity in vitro and in vivo.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


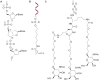


Similar articles
-
Designing Effective Antisense Oligonucleotides for Exon Skipping.Methods Mol Biol. 2018;1687:143-155. doi: 10.1007/978-1-4939-7374-3_10. Methods Mol Biol. 2018. PMID: 29067661
-
New developments in exon skipping and splice modulation therapies for neuromuscular diseases.Expert Opin Biol Ther. 2014 Jun;14(6):809-19. doi: 10.1517/14712598.2014.896335. Epub 2014 Mar 12. Expert Opin Biol Ther. 2014. PMID: 24620745 Review.
-
Systematic evaluation of 2'-Fluoro modified chimeric antisense oligonucleotide-mediated exon skipping in vitro.Sci Rep. 2019 Apr 15;9(1):6078. doi: 10.1038/s41598-019-42523-0. Sci Rep. 2019. PMID: 30988454 Free PMC article.
-
Genetic neuromuscular disorders: living the era of a therapeutic revolution. Part 2: diseases of motor neuron and skeletal muscle.Neurol Sci. 2019 Apr;40(4):671-681. doi: 10.1007/s10072-019-03764-z. Epub 2019 Feb 25. Neurol Sci. 2019. PMID: 30805745 Review.
-
Short (16-mer) locked nucleic acid splice-switching oligonucleotides restore dystrophin production in Duchenne Muscular Dystrophy myotubes.PLoS One. 2017 Jul 24;12(7):e0181065. doi: 10.1371/journal.pone.0181065. eCollection 2017. PLoS One. 2017. PMID: 28742140 Free PMC article.
Cited by
-
Splice-Modulating Antisense Oligonucleotides as Therapeutics for Inherited Metabolic Diseases.BioDrugs. 2024 Mar;38(2):177-203. doi: 10.1007/s40259-024-00644-7. Epub 2024 Jan 22. BioDrugs. 2024. PMID: 38252341 Free PMC article. Review.
-
Introduction to the themed collection in honour of Professor Christian Leumann.RSC Med Chem. 2024 Oct 22;15(11):3636-8. doi: 10.1039/d4md90039a. Online ahead of print. RSC Med Chem. 2024. PMID: 39450116 Free PMC article.
-
Evaluation of Chemically Modified Nucleic Acid Analogues for Splice Switching Application.ACS Omega. 2023 Dec 11;8(51):48650-48661. doi: 10.1021/acsomega.3c07618. eCollection 2023 Dec 26. ACS Omega. 2023. PMID: 38162739 Free PMC article. Review.
-
A Visual Compendium of Principal Modifications within the Nucleic Acid Sugar Phosphate Backbone.Molecules. 2024 Jun 26;29(13):3025. doi: 10.3390/molecules29133025. Molecules. 2024. PMID: 38998973 Free PMC article. Review.
-
Amplifying gene expression with RNA-targeted therapeutics.Nat Rev Drug Discov. 2023 Jul;22(7):539-561. doi: 10.1038/s41573-023-00704-7. Epub 2023 May 30. Nat Rev Drug Discov. 2023. PMID: 37253858 Free PMC article. Review.
References
-
- Morvan F., Rayner B., Imbach J.L., Thenet S., Bertrand J.R., Paoletti J., Malvy C., Paoletti C.. alpha-DNA II. Synthesis of unnatural alpha-anomeric oligodeoxyribonucleotides containing the four usual bases and study of their substrate activities for nucleases. Nucleic Acids Res. 1987; 15:3421–3437. - PMC - PubMed
-
- Morvan F., Rayner B., Imbach J.L., Chang D.K., Lown J.W.. alpha-DNA-III. Characterization by high field 1H-NMR, anti-parallel self-recognition and conformation of the unnatural hexadeoxyribonucleotides alpha-[d(CpApTpGpCpG)] and alpha-[d(CpGpCpApTpG)]. Alpha-oligodeoxynucleotides as potential cellular probes for gene control. Nucleic Acids Res. 1987; 15:4241–4255. - PMC - PubMed
-
- Morvan F., Rayner B., Imbach J.L., Chang D.K., Lown J.W.. alpha-DNA. I. Synthesis, characterization by high field 1H-NMR, and base-pairing properties of the unnatural hexadeoxyribonucleotide alpha-[d(CpCpTpTpCpC)] with its complement beta-[d(GpGpApApGpG)]. Nucleic Acids Res. 1986; 14:5019–5035. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

