Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases
- PMID: 34850143
- PMCID: PMC8643638
- DOI: 10.1093/nar/gkab1130
Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases
Abstract
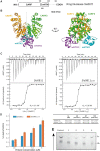
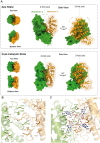
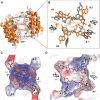
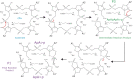
Type III CRISPR-Cas effector systems detect foreign RNA triggering DNA and RNA cleavage and synthesizing cyclic oligoadenylate molecules (cA) in their Cas10 subunit. cAs act as a second messenger activating auxiliary nucleases, leading to an indiscriminate RNA degradation that can end in cell dormancy or death. Standalone ring nucleases are CRISPR ancillary proteins which downregulate the strong immune response of Type III systems by degrading cA. These enzymes contain a CRISPR-associated Rossman-fold (CARF) domain, which binds and cleaves the cA molecule. Here, we present the structures of the standalone ring nuclease from Sulfolobus islandicus (Sis) 0811 in its apo and post-catalytic states. This enzyme is composed by a N-terminal CARF and a C-terminal wHTH domain. Sis0811 presents a phosphodiester hydrolysis metal-independent mechanism, which cleaves cA4 rings to generate linear adenylate species, thus reducing the levels of the second messenger and switching off the cell antiviral state. The structural and biochemical analysis revealed the coupling of a cork-screw conformational change with the positioning of key catalytic residues to proceed with cA4 phosphodiester hydrolysis in a non-concerted manner.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Molecular basis of cyclic tetra-oligoadenylate processing by small standalone CRISPR-Cas ring nucleases.Nucleic Acids Res. 2022 Oct 28;50(19):11199-11213. doi: 10.1093/nar/gkac923. Nucleic Acids Res. 2022. PMID: 36271789 Free PMC article.
-
Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate.Nature. 2018 Oct;562(7726):277-280. doi: 10.1038/s41586-018-0557-5. Epub 2018 Sep 19. Nature. 2018. PMID: 30232454 Free PMC article.
-
Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6.Nat Commun. 2020 Mar 27;11(1):1596. doi: 10.1038/s41467-020-15334-5. Nat Commun. 2020. PMID: 32221291 Free PMC article.
-
Type III CRISPR-Cas: beyond the Cas10 effector complex.Trends Biochem Sci. 2024 Jan;49(1):28-37. doi: 10.1016/j.tibs.2023.10.006. Epub 2023 Nov 8. Trends Biochem Sci. 2024. PMID: 37949766 Free PMC article. Review.
-
The diverse arsenal of type III CRISPR-Cas-associated CARF and SAVED effectors.Biochem Soc Trans. 2022 Oct 31;50(5):1353-1364. doi: 10.1042/BST20220289. Biochem Soc Trans. 2022. PMID: 36282000 Free PMC article. Review.
Cited by
-
The SAVED domain of the type III CRISPR protease CalpL is a ring nuclease.Nucleic Acids Res. 2024 Sep 23;52(17):10520-10532. doi: 10.1093/nar/gkae676. Nucleic Acids Res. 2024. PMID: 39166476 Free PMC article.
-
Molecular basis of stepwise cyclic tetra-adenylate cleavage by the type III CRISPR ring nuclease Crn1/Sso2081.Nucleic Acids Res. 2023 Mar 21;51(5):2485-2495. doi: 10.1093/nar/gkad101. Nucleic Acids Res. 2023. PMID: 36807980 Free PMC article.
-
Molecular mechanism of allosteric activation of the CRISPR ribonuclease Csm6 by cyclic tetra-adenylate.EMBO J. 2024 Jan;43(2):304-315. doi: 10.1038/s44318-023-00017-w. Epub 2023 Dec 19. EMBO J. 2024. PMID: 38177499 Free PMC article.
-
An archaeal virus-encoded anti-CRISPR protein inhibits type III-B immunity by inhibiting Cas RNP complex turnover.Nucleic Acids Res. 2023 Nov 27;51(21):11783-11796. doi: 10.1093/nar/gkad804. Nucleic Acids Res. 2023. PMID: 37850639 Free PMC article.
-
Knowing Our Enemy in the Antimicrobial Resistance Era: Dissecting the Molecular Basis of Bacterial Defense Systems.Int J Mol Sci. 2024 Apr 30;25(9):4929. doi: 10.3390/ijms25094929. Int J Mol Sci. 2024. PMID: 38732145 Free PMC article. Review.
References
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709. - PubMed
-
- Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.v, van der Oost J.. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016; 353:aad5147. - PubMed
-
- Amitai G., Sorek R.. CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016; 14:67–76. - PubMed
-
- Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.J.. CRISPR-Cas: adapting to change. Science. 2017; 356:eaal5056. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

