Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly
- PMID: 34850144
- PMCID: PMC8682754
- DOI: 10.1093/nar/gkab1155
Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly
Abstract
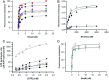
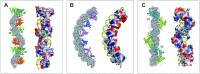
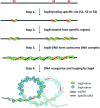
Genome segregation is a vital process in all organisms. Chromosome partitioning remains obscure in Archaea, the third domain of life. Here, we investigated the SegAB system from Sulfolobus solfataricus. SegA is a ParA Walker-type ATPase and SegB is a site-specific DNA-binding protein. We determined the structures of both proteins and those of SegA-DNA and SegB-DNA complexes. The SegA structure revealed an atypical, novel non-sandwich dimer that binds DNA either in the presence or in the absence of ATP. The SegB structure disclosed a ribbon-helix-helix motif through which the protein binds DNA site specifically. The association of multiple interacting SegB dimers with the DNA results in a higher order chromatin-like structure. The unstructured SegB N-terminus plays an essential catalytic role in stimulating SegA ATPase activity and an architectural regulatory role in segrosome (SegA-SegB-DNA) formation. Electron microscopy results also provide a compact ring-like segrosome structure related to chromosome organization. These findings contribute a novel mechanistic perspective on archaeal chromosome segregation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Hayes F., Barilla D.. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006; 4:133–143. - PubMed
-
- Prosser S.L., Pelletier L.. Mitotic spindle assembly in animal cells: a fine balancing act. Nat. Rev. Mol. Cell Biol. 2017; 18:187–201. - PubMed
-
- Hurtgen D., Murray S.M., Mascarenhas J., Sourjik V.. DNA segregation in natural and synthetic minimal systems. Adv. Biosyst. 2019; 3:e1800316. - PubMed
-
- Baxter J.C., Funnell B.E.. Plasmid partition mechanisms. Microbiol. Spectr. 2014; 2:6. - PubMed
-
- Gerdes K., Howard M., Szardenings F.. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010; 141:927–942. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

